Characterization of a central Ca2+/calmodulin-dependent protein kinase IIalpha/beta binding domain in densin that selectively modulates glutamate receptor subunit phosphorylation
- PMID: 21610080
- PMCID: PMC3137056
- DOI: 10.1074/jbc.M110.216010
Characterization of a central Ca2+/calmodulin-dependent protein kinase IIalpha/beta binding domain in densin that selectively modulates glutamate receptor subunit phosphorylation
Abstract
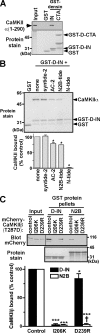
The densin C-terminal domain can target Ca(2+)/calmodulin-dependent protein kinase IIα (CaMKIIα) in cells. Although the C-terminal domain selectively binds CaMKIIα in vitro, full-length densin associates with CaMKIIα or CaMKIIβ in brain extracts and in transfected HEK293 cells. This interaction requires a second central CaMKII binding site, the densin-IN domain, and an "open" activated CaMKII conformation caused by Ca(2+)/calmodulin binding, autophosphorylation at Thr-286/287, or mutation of Thr-286/287 to Asp. Mutations in the densin-IN domain (L815E) or in the CaMKIIα/β catalytic domain (I205/206K) disrupt the interaction. The amino acid sequence of the densin-IN domain is similar to the CaMKII inhibitor protein, CaMKIIN, and a CaMKIIN peptide competitively blocks CaMKII binding to densin. CaMKII is inhibited by both CaMKIIN and the densin-IN domain, but the inhibition by densin is substrate-selective. Phosphorylation of a model peptide substrate, syntide-2, or of Ser-831 in AMPA receptor GluA1 subunits is fully inhibited by densin. However, CaMKII phosphorylation of Ser-1303 in NMDA receptor GluN2B subunits is not effectively inhibited by densin in vitro or in intact cells. Thus, densin can target multiple CaMKII isoforms to differentially modulate phosphorylation of physiologically relevant downstream targets.
Figures







Similar articles
-
Differential modulation of Ca2+/calmodulin-dependent protein kinase II activity by regulated interactions with N-methyl-D-aspartate receptor NR2B subunits and alpha-actinin.J Biol Chem. 2005 Nov 25;280(47):39316-23. doi: 10.1074/jbc.M508189200. Epub 2005 Sep 19. J Biol Chem. 2005. PMID: 16172120
-
Multivalent interactions of calcium/calmodulin-dependent protein kinase II with the postsynaptic density proteins NR2B, densin-180, and alpha-actinin-2.J Biol Chem. 2005 Oct 21;280(42):35329-36. doi: 10.1074/jbc.M502191200. Epub 2005 Aug 24. J Biol Chem. 2005. PMID: 16120608
-
Densin-180 forms a ternary complex with the (alpha)-subunit of Ca2+/calmodulin-dependent protein kinase II and (alpha)-actinin.J Neurosci. 2001 Jan 15;21(2):423-33. doi: 10.1523/JNEUROSCI.21-02-00423.2001. J Neurosci. 2001. PMID: 11160423 Free PMC article.
-
Structural Insights into the Regulation of Ca2+/Calmodulin-Dependent Protein Kinase II (CaMKII).Cold Spring Harb Perspect Biol. 2020 Jun 1;12(6):a035147. doi: 10.1101/cshperspect.a035147. Cold Spring Harb Perspect Biol. 2020. PMID: 31653643 Free PMC article. Review.
-
Postsynaptic localization and regulation of AMPA receptors and Cav1.2 by β2 adrenergic receptor/PKA and Ca2+/CaMKII signaling.EMBO J. 2018 Oct 15;37(20):e99771. doi: 10.15252/embj.201899771. Epub 2018 Sep 24. EMBO J. 2018. PMID: 30249603 Free PMC article. Review.
Cited by
-
CaMKII regulation in information processing and storage.Trends Neurosci. 2012 Oct;35(10):607-18. doi: 10.1016/j.tins.2012.05.003. Epub 2012 Jun 19. Trends Neurosci. 2012. PMID: 22717267 Free PMC article. Review.
-
A frontier in the understanding of synaptic plasticity: solving the structure of the postsynaptic density.Bioessays. 2012 Jul;34(7):599-608. doi: 10.1002/bies.201200009. Epub 2012 Apr 23. Bioessays. 2012. PMID: 22528972 Free PMC article. Review.
-
CaMKII inhibitors: from research tools to therapeutic agents.Front Pharmacol. 2014 Feb 20;5:21. doi: 10.3389/fphar.2014.00021. eCollection 2014. Front Pharmacol. 2014. PMID: 24600394 Free PMC article. Review.
-
Variants in LRRC7 lead to intellectual disability, autism, aggression and abnormal eating behaviors.Nat Commun. 2024 Sep 10;15(1):7909. doi: 10.1038/s41467-024-52095-x. Nat Commun. 2024. PMID: 39256359 Free PMC article.
-
Activity dependent Clustering of Neuronal L-Type Calcium Channels by CaMKII.bioRxiv [Preprint]. 2025 Jan 8:2025.01.08.631979. doi: 10.1101/2025.01.08.631979. bioRxiv. 2025. PMID: 39829809 Free PMC article. Preprint.
References
-
- Barria A., Malinow R. (2005) Neuron 48, 289–301 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

