A novel methodology for large-scale phylogeny partition
- PMID: 21610724
- PMCID: PMC6045912
- DOI: 10.1038/ncomms1325
A novel methodology for large-scale phylogeny partition
Abstract
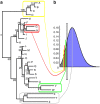
Understanding the determinants of virus transmission is a fundamental step for effective design of screening and intervention strategies to control viral epidemics. Phylogenetic analysis can be a valid approach for the identification of transmission chains, and very-large data sets can be analysed through parallel computation. Here we propose and validate a new methodology for the partition of large-scale phylogenies and the inference of transmission clusters. This approach, on the basis of a depth-first search algorithm, conjugates the evaluation of node reliability, tree topology and patristic distance analysis. The method has been applied to identify transmission clusters of a phylogeny of 11,541 human immunodeficiency virus-1 subtype B pol gene sequences from a large Italian cohort. Molecular transmission chains were characterized by means of different clinical/demographic factors, such as the interaction between male homosexuals and male heterosexuals. Our method takes an advantage of a flexible notion of transmission cluster and can become a general framework to analyse other epidemics.
© 2011 Macmillan Publishers Limited. All rights reserved.
Conflict of interest statement
A.De L. received speakers' honoraria, served as consultant or participated in advisory boards for GlaxoSmithKline, Gilead, Bristol-Myers Squibb, Abbott Virology, Janssen-Cilag Tibotec, Siemens Diagnostics and Monogram Biosciences. S.R. has received research grants and has been involved in advisory boards or educational courses supported by the following companies: Abbott, Boehringer-Ingelheim, Bristol-Myers Squibb, Gilead, GlaxoSmithKline, ViiV Healthcare, Merck, and Janssen-Cilag. M.Z. has received research funding from Pfizer; served as a consultant for Abbott Molecular, Boehringer Ingelheim, Gilead Sciences and Janssen-Cilag; and served on speakers' bureaus for Abbott, Bristol-Myers Squibb, Merck, and Pfizer. The remaining authors declare no competing financial interests. B.B. has received funds for speaking, consultancy and travel from ViiV Healthcare, Gilead Sciences, Abbott Molecular, Janssen-Cilag and Siemens Health Care.
Figures