Epigenetically silenced miR-34b/c as a novel faecal-based screening marker for colorectal cancer
- PMID: 21610744
- PMCID: PMC3111174
- DOI: 10.1038/bjc.2011.82
Epigenetically silenced miR-34b/c as a novel faecal-based screening marker for colorectal cancer
Abstract
Background: MicroRNAs are tiny non-coding small endogenous RNAs that regulate gene expression by translational repression, mRNA cleavage and mRNA inhibition. The aim of this study was to investigate the hypermethylation of miR-34b/c and miR-148a in colorectal cancer, and correlate this data to clinicopathological features. We also aimed to evaluate the hypermethylation of miR-34b/c in faeces specimens as a novel non-invasive faecal-DNA-based screening marker.
Methods: The 5-aza-2'-deoxycytidine treatment and methylation-specific PCR were carried out to detect the hypermethylation of miR-34b/c and miR-148a.
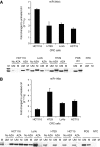
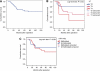
Results: The miR-34b/c hypermethylation was found in 97.5% (79 out of 82) of primary colorectal tumours, P=0.0110. In 75% (21 out of 28) of faecal specimens we found a hypermethylation of miR-34b/c while only in 16% (2 out of 12) of high-grade dysplasia. In addition, miR-148a was found to be hypermethylated in 65% (51 out of 78) of colorectal tumour tissues with no significant correlation to clinicopathological features. However, a trend with female gender and advanced age was found, P=0.083. We also observed a trend to lower survival rate in patients with miR-148a hypermethylation with 10-year survival probability: 48 vs 65%, P=0.561.
Conclusions: These findings show that aberrant hypermethylation of miR-34b/c could be an ideal class of early screening marker, whereas miR-148a could serve as a disease progression follow-up marker.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Epigenetic alteration of microRNAs in feces of colorectal cancer and its clinical significance.Expert Rev Mol Diagn. 2011 Sep;11(7):691-4. doi: 10.1586/erm.11.57. Expert Rev Mol Diagn. 2011. PMID: 21902530
References
-
- Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, Prosper F, Garcia-Foncillas J (2009) Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer 125: 2737–2743 - PubMed
-
- Barbarotto E, Schmittgen TD, Calin GA (2008) MicroRNAs andcancer: profile, profile, profile. Int J Cancer 122: 969–977 - PubMed
-
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

