Treatment of spinal muscular atrophy cells with drugs that upregulate SMN expression reveals inter- and intra-patient variability
- PMID: 21610752
- PMCID: PMC3190259
- DOI: 10.1038/ejhg.2011.89
Treatment of spinal muscular atrophy cells with drugs that upregulate SMN expression reveals inter- and intra-patient variability
Abstract
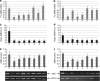
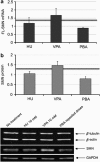
Spinal muscular atrophy (SMA) is a genetic neuromuscular disorder caused by mutations in the SMN1 gene. The homologous copy (SMN2) is always present in SMA patients. SMN1 gene transcripts are usually full-length (FL), but exon 7 is spliced out in a high proportion of SMN2 transcripts (delta7) (Δ7). Advances in drug therapy for SMA have shown that an increase in SMN mRNA and protein levels can be achieved in vitro. We performed a systematic analysis of SMN expression in primary fibroblasts and EBV-transformed lymphoblasts from seven SMA patients with varying clinical severity and different SMN1 genotypes to determine expression differences in two accessible tissues (skin and blood). The basal expression of SMN mRNA FL and Δ7 in fibroblasts and lymphoblasts was analyzed by quantitative real-time PCR. The FL-SMN and FL/Δ7 SMN ratios were higher in control cells than in patients. Furthermore, we investigated the response of these cell lines to hydroxyurea, valproate and phenylbutyrate, drugs previously reported to upregulate SMN2. The response to treatments with these compounds was heterogeneous. We found both intra-patient and inter-patient variability even within haploidentical siblings, suggesting that tissue and individual factors may affect the response to these compounds. To optimize the stratification of patients in clinical trials, in vitro studies should be performed before enrolment so as to define each patient as a responder or non-responder to the compound under investigation.
Figures



References
-
- Alias L, Bernal S, Fuentes-Prior P, et al. Mutation update of spinal muscular atrophy in Spain: molecular characterization of 745 unrelated patients and identification of four novel mutations in the SMN1 gene. Hum Genet. 2009;125:29–39. - PubMed
-
- Lefebvre S, Burglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. - PubMed
-
- Bussaglia E, Clermont O, Tizzano E, et al. A frame-shift deletion in the survival motor neuron gene in Spanish spinal muscular atrophy patients. Nat Genet. 1995;11:335–337. - PubMed
-
- Cusco I, Lopez E, Soler-Botija C, Jesus Barcelo M, Baiget M, Tizzano EF. A genetic and phenotypic analysis in Spanish spinal muscular atrophy patients with c.399_402del AGAG, the most frequently found subtle mutation in the SMN1 gene. Hum Mutat. 2003;22:136–143. - PubMed
-
- Martin Y, Valero A, del Castillo E, Pascual SI, Hernandez-Chico C. Genetic study of SMA patients without homozygous SMN1 deletions: identification of compound heterozygotes and characterisation of novel intragenic SMN1 mutations. Hum Genet. 2002;110:257–263. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

