Orientia tsutsugamushi stimulates an original gene expression program in monocytes: relationship with gene expression in patients with scrub typhus
- PMID: 21610853
- PMCID: PMC3096591
- DOI: 10.1371/journal.pntd.0001028
Orientia tsutsugamushi stimulates an original gene expression program in monocytes: relationship with gene expression in patients with scrub typhus
Expression of concern in
-
Expression of Concern: Orientia tsutsugamushi Stimulates an Original Gene Expression Program in Monocytes: Relationship with Gene Expression in Patients with Scrub Typhus.PLoS Negl Trop Dis. 2022 Dec 13;16(12):e0010958. doi: 10.1371/journal.pntd.0010958. eCollection 2022 Dec. PLoS Negl Trop Dis. 2022. PMID: 36512535 Free PMC article. No abstract available.
Abstract
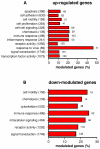
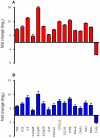
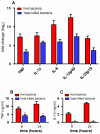
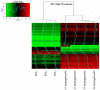
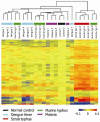
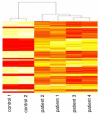
Orientia tsutsugamushi is the causal agent of scrub typhus, a public health problem in the Asia-Pacific region and a life-threatening disease. O. tsutsugamushi is an obligate intracellular bacterium that mainly infects endothelial cells. We demonstrated here that O. tsutsugamushi also replicated in monocytes isolated from healthy donors. In addition, O. tsutsugamushi altered the expression of more than 4,500 genes, as demonstrated by microarray analysis. The expression of type I interferon, interferon-stimulated genes and genes associated with the M1 polarization of macrophages was significantly upregulated. O. tsutsugamushi also induced the expression of apoptosis-related genes and promoted cell death in a small percentage of monocytes. Live organisms were indispensable to the type I interferon response and apoptosis and enhanced the expression of M1-associated cytokines. These data were related to the transcriptional changes detected in mononuclear cells isolated from patients with scrub typhus. Here, the microarray analyses revealed the upregulation of 613 genes, which included interferon-related genes, and some features of M1 polarization were observed in these patients, similar to what was observed in O. tsutsugamushi-stimulated monocytes in vitro. This is the first report demonstrating that monocytes are clearly polarized in vitro and ex vivo following exposure to O. tsutsugamushi. These results would improve our understanding of the pathogenesis of scrub typhus, during which interferon-mediated activation of monocytes and their subsequent polarization into an M1 phenotype appear critical. This study may give us a clue of new tools for the diagnosis of patients with scrub typhus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Seong SY, Choi MS, Kim IS. Orientia tsutsugamushi infection: overview and immune responses. Microbes Infect. 2001;3:11–21. - PubMed
-
- Watt G, Parola P. Scrub typhus and tropical rickettsioses. Curr Opin Infect Dis. 2003;16:429–436. - PubMed
-
- Chattopadhyay S, Richards AL. Scrub typhus vaccines: past history and recent developments. Hum Vaccin. 2007;3:73–80. - PubMed
-
- Tamura A, Ohashi N, Urakami U, Miyamura S. Classification of Rickettsia tsutsugamushi in a new genus, Orientia gen. nov., as Orientia tsutsugamushi comb. nov. Int J Syst Bacteriol. 1995;45:589–591. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

