Toward a DNA taxonomy of Alpine Rhithrogena (Ephemeroptera: Heptageniidae) using a mixed Yule-coalescent analysis of mitochondrial and nuclear DNA
- PMID: 21611178
- PMCID: PMC3096624
- DOI: 10.1371/journal.pone.0019728
Toward a DNA taxonomy of Alpine Rhithrogena (Ephemeroptera: Heptageniidae) using a mixed Yule-coalescent analysis of mitochondrial and nuclear DNA
Abstract
Aquatic larvae of many Rhithrogena mayflies (Ephemeroptera) inhabit sensitive Alpine environments. A number of species are on the IUCN Red List and many recognized species have restricted distributions and are of conservation interest. Despite their ecological and conservation importance, ambiguous morphological differences among closely related species suggest that the current taxonomy may not accurately reflect the evolutionary diversity of the group. Here we examined the species status of nearly 50% of European Rhithrogena diversity using a widespread sampling scheme of Alpine species that included 22 type localities, general mixed Yule-coalescent (GMYC) model analysis of one standard mtDNA marker and one newly developed nDNA marker, and morphological identification where possible. Using sequences from 533 individuals from 144 sampling localities, we observed significant clustering of the mitochondrial (cox1) marker into 31 GMYC species. Twenty-one of these could be identified based on the presence of topotypes (expertly identified specimens from the species' type locality) or unambiguous morphology. These results strongly suggest the presence of both cryptic diversity and taxonomic oversplitting in Rhithrogena. Significant clustering was not detected with protein-coding nuclear PEPCK, although nine GMYC species were congruent with well supported terminal clusters of nDNA. Lack of greater congruence in the two data sets may be the result of incomplete sorting of ancestral polymorphism. Bayesian phylogenetic analyses of both gene regions recovered four of the six recognized Rhithrogena species groups in our samples as monophyletic. Future development of more nuclear markers would facilitate multi-locus analysis of unresolved, closely related species pairs. The DNA taxonomy developed here lays the groundwork for a future revision of the important but cryptic Rhithrogena genus in Europe.
Conflict of interest statement
Figures

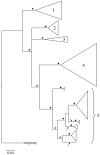


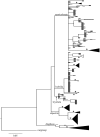
References
-
- Petit RJ, Excoffier L. Gene flow and species delimitation. Trends in Ecology and Evolution. 2009;24:386–393. - PubMed
-
- Sites JW, Marshall JC. Delimiting species: a Renaissance issue in systematic biology. Trends in Ecology & Evolution 2003
-
- Knowles LL, Carstens BC. Delimiting species without monophyletic gene trees. Systematic Biology. 2007;56:887–895. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

