Regulation of flowering time: all roads lead to Rome
- PMID: 21611891
- PMCID: PMC11115107
- DOI: 10.1007/s00018-011-0673-y
Regulation of flowering time: all roads lead to Rome
Abstract
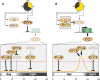
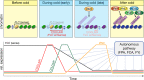
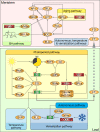
Plants undergo a major physiological change as they transition from vegetative growth to reproductive development. This transition is a result of responses to various endogenous and exogenous signals that later integrate to result in flowering. Five genetically defined pathways have been identified that control flowering. The vernalization pathway refers to the acceleration of flowering on exposure to a long period of cold. The photoperiod pathway refers to regulation of flowering in response to day length and quality of light perceived. The gibberellin pathway refers to the requirement of gibberellic acid for normal flowering patterns. The autonomous pathway refers to endogenous regulators that are independent of the photoperiod and gibberellin pathways. Most recently, an endogenous pathway that adds plant age to the control of flowering time has been described. The molecular mechanisms of these pathways have been studied extensively in Arabidopsis thaliana and several other flowering plants.
Figures
References
-
- Kobayashi Y, Weigel D. Move on up, it’s time for change—mobile signals controlling photoperiod-dependent flowering. Genes Dev. 2007;21:2371–2384. - PubMed
-
- Pittendrigh CS. Circadian rhythms and the circadian organization of living systems. Cold Spring Harb Symp Quant Biol. 1960;25:159–184. - PubMed
-
- Knott J. Effect of a localized photoperiod on spinach. Proc Soc Hortic Sci. 1934;31:152–154.
-
- Evans LT. Flower induction and the florigen concept. Annu Rev Plant Physiol. 1971;22:365–394.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

