The crystal structure of alanine racemase from Streptococcus pneumoniae, a target for structure-based drug design
- PMID: 21612658
- PMCID: PMC3146814
- DOI: 10.1186/1471-2180-11-116
The crystal structure of alanine racemase from Streptococcus pneumoniae, a target for structure-based drug design
Abstract
Background: Streptococcus pneumoniae is a globally important pathogen. The Gram-positive diplococcus is a leading cause of pneumonia, otitis media, bacteremia, and meningitis, and antibiotic resistant strains have become increasingly common over recent years. Alanine racemase is a ubiquitous enzyme among bacteria and provides the essential cell wall precursor, D-alanine. Since it is absent in humans, this enzyme is an attractive target for the development of drugs against S. pneumoniae and other bacterial pathogens.
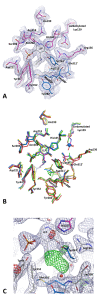
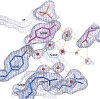
Results: Here we report the crystal structure of alanine racemase from S. pneumoniae (AlrSP). Crystals diffracted to a resolution of 2.0 Å and belong to the space group P3121 with the unit cell parameters a = b = 119.97 Å, c = 118.10 Å, α = β = 90° and γ = 120°. Structural comparisons show that AlrSP shares both an overall fold and key active site residues with other bacterial alanine racemases. The active site cavity is similar to other Gram positive alanine racemases, featuring a restricted but conserved entryway.
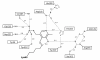
Conclusions: We have solved the structure of AlrSP, an essential step towards the development of an accurate pharmacophore model of the enzyme, and an important contribution towards our on-going alanine racemase structure-based drug design project. We have identified three regions on the enzyme that could be targeted for inhibitor design, the active site, the dimer interface, and the active site entryway.
Figures


Similar articles
-
Purification and preliminary crystallization of alanine racemase from Streptococcus pneumoniae.BMC Microbiol. 2007 May 17;7:40. doi: 10.1186/1471-2180-7-40. BMC Microbiol. 2007. PMID: 17509154 Free PMC article.
-
Structural features and kinetic characterization of alanine racemase from Staphylococcus aureus (Mu50).Acta Crystallogr D Biol Crystallogr. 2012 Jan;68(Pt 1):82-92. doi: 10.1107/S0907444911050682. Epub 2011 Dec 9. Acta Crystallogr D Biol Crystallogr. 2012. PMID: 22194336 Free PMC article.
-
The structure of alanine racemase from Acinetobacter baumannii.Acta Crystallogr F Struct Biol Commun. 2014 Sep;70(Pt 9):1199-205. doi: 10.1107/S2053230X14017725. Epub 2014 Aug 29. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 25195891 Free PMC article.
-
Inhibitors of alanine racemase enzyme: a review.J Enzyme Inhib Med Chem. 2016 Aug;31(4):517-26. doi: 10.3109/14756366.2015.1050010. Epub 2015 May 29. J Enzyme Inhib Med Chem. 2016. PMID: 26024289 Review.
-
Threonine aldolase and alanine racemase: novel examples of convergent evolution in the superfamily of vitamin B6-dependent enzymes.Biochim Biophys Acta. 2003 Apr 11;1647(1-2):214-9. doi: 10.1016/s1570-9639(03)00050-5. Biochim Biophys Acta. 2003. PMID: 12686135 Review.
Cited by
-
Crystal Structure of a Thermostable Alanine Racemase from Thermoanaerobacter tengcongensis MB4 Reveals the Role of Gln360 in Substrate Selection.PLoS One. 2015 Jul 28;10(7):e0133516. doi: 10.1371/journal.pone.0133516. eCollection 2015. PLoS One. 2015. PMID: 26218070 Free PMC article.
-
Purification, Characterization and Inhibition of Alanine Racemase from a Pathogenic Strain of Streptococcus iniae.Pol J Microbiol. 2019 Sep;68(3):331-341. doi: 10.33073/pjm-2019-036. Epub 2019 Sep 3. Pol J Microbiol. 2019. PMID: 31880879 Free PMC article.
-
Selection and characterization of alanine racemase inhibitors against Aeromonas hydrophila.BMC Microbiol. 2017 May 25;17(1):122. doi: 10.1186/s12866-017-1010-x. BMC Microbiol. 2017. PMID: 28545531 Free PMC article.
-
In Silico Identification and Characterization of Drug Targets in Streptococcus pneumoniae ATCC 700669 (Serotype 23F) by Subtractive Genomics.Biomed Res Int. 2024 Jan 20;2024:5917667. doi: 10.1155/2024/5917667. eCollection 2024. Biomed Res Int. 2024. PMID: 38283072 Free PMC article.
-
Crystallization and preliminary X-ray study of a thermostable alanine racemase from Thermoanaerobacter tengcongensis MB4.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 Jun;69(Pt 6):660-2. doi: 10.1107/S1744309113011743. Epub 2013 May 25. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 23722847 Free PMC article.
References
-
- Osler SW. Aequanimitas: with other addresses to medical students, nurses and and practitioners of medicine. Philadelphia: P. Blakiston's Son & Co; 1905. Medicine in the Nineteenth Century; pp. 217–262.
-
- World Health Organization Initiative for Vaccine Research. Acute Respiratory Infections (Update September 2009) http://www.who.int/vaccine_research/diseases/ari/en/
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

