Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus
- PMID: 21613567
- PMCID: PMC3116389
- DOI: 10.1073/pnas.1012388108
Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus
Erratum in
- Proc Natl Acad Sci U S A. 2011 Jul 19;108(29):12185
Abstract
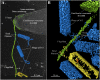
2D and 3D cryo-electron microscopy, together with adsorption kinetics assays of Cb13 and CbK phage-infected Caulobacter crescentus, provides insight into the mechanisms of infection. Cb13 and CbK actively interact with the flagellum and subsequently attach to receptors on the cell pole. We present evidence that the first interaction of the phage with the bacterial flagellum takes place through a filament on the phage head. This contact with the flagellum facilitates concentration of phage particles around the receptor (i.e., the pilus portals) on the bacterial cell surface, thereby increasing the likelihood of infection. Phage head filaments have not been well characterized and their function is described here. Phage head filaments may systematically underlie the initial interactions of phages with their hosts in other systems and possibly represent a widespread mechanism of efficient phage propagation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

