Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis
- PMID: 21613568
- PMCID: PMC3116379
- DOI: 10.1073/pnas.1103584108
Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis
Abstract
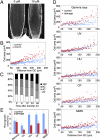
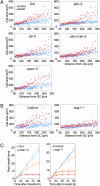
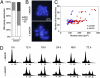
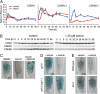
Genome integrity is continuously threatened by external stresses and endogenous hazards such as DNA replication errors and reactive oxygen species. The DNA damage checkpoint in metazoans ensures genome integrity by delaying cell-cycle progression to repair damaged DNA or by inducing apoptosis. ATM and ATR (ataxia-telangiectasia-mutated and -Rad3-related) are sensor kinases that relay the damage signal to transducer kinases Chk1 and Chk2 and to downstream cell-cycle regulators. Plants also possess ATM and ATR orthologs but lack obvious counterparts of downstream regulators. Instead, the plant-specific transcription factor SOG1 (suppressor of gamma response 1) plays a central role in the transmission of signals from both ATM and ATR kinases. Here we show that in Arabidopsis, endoreduplication is induced by DNA double-strand breaks (DSBs), but not directly by DNA replication stress. When root or sepal cells, or undifferentiated suspension cells, were treated with DSB inducers, they displayed increased cell size and DNA ploidy. We found that the ATM-SOG1 and ATR-SOG1 pathways both transmit DSB-derived signals and that either one suffices for endocycle induction. These signaling pathways govern the expression of distinct sets of cell-cycle regulators, such as cyclin-dependent kinases and their suppressors. Our results demonstrate that Arabidopsis undergoes a programmed endoreduplicative response to DSBs, suggesting that plants have evolved a distinct strategy to sustain growth under genotoxic stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. - PubMed
-
- Rotman G, Shiloh Y. ATM: From gene to function. Hum Mol Genet. 1998;7:1555–1563. - PubMed
-
- de Klein A, et al. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000;10:479–482. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

