An incoherent regulatory network architecture that orchestrates B cell diversification in response to antigen signaling
- PMID: 21613984
- PMCID: PMC3130558
- DOI: 10.1038/msb.2011.25
An incoherent regulatory network architecture that orchestrates B cell diversification in response to antigen signaling
Abstract
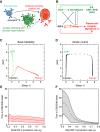
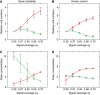
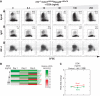
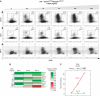
The B-lymphocyte lineage is a leading system for analyzing gene regulatory networks (GRNs) that orchestrate distinct cell fate transitions. Upon antigen recognition, B cells can diversify their immunoglobulin (Ig) repertoire via somatic hypermutation (SHM) and/or class switch DNA recombination (CSR) before differentiating into antibody-secreting plasma cells. We construct a mathematical model for a GRN underlying this developmental dynamic. The intensity of signaling through the Ig receptor is shown to control the bimodal expression of a pivotal transcription factor, IRF-4, which dictates B cell fate outcomes. Computational modeling coupled with experimental analysis supports a model of 'kinetic control', in which B cell developmental trajectories pass through an obligate transient state of variable duration that promotes diversification of the antibody repertoire by SHM/CSR in direct response to antigens. More generally, this network motif could be used to translate a morphogen gradient into developmental inductive events of varying time, thereby enabling the specification of distinct cell fates.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Acar M, Becskei A, van Oudenaarden A (2005) Enhancement of cellular memory by reducing stochastic transitions. Nature 435: 228–232 - PubMed
-
- Alon U (2007) An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton: Chapman & Hall/CRC
-
- Aurell E, Brown S, Johanson J, Sneppen K (2002) Stability puzzles in phage λ. Phys Rev E Stat Nonlin Soft Matter Phys 65: 051914. - PubMed
-
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R (2006) Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44: 23–28 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

