Genes that escape from X inactivation
- PMID: 21614513
- PMCID: PMC3136209
- DOI: 10.1007/s00439-011-1011-z
Genes that escape from X inactivation
Abstract
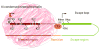
To achieve a balanced gene expression dosage between males (XY) and females (XX), mammals have evolved a compensatory mechanism to randomly inactivate one of the female X chromosomes. Despite this chromosome-wide silencing, a number of genes escape X inactivation: in women about 15% of X-linked genes are bi-allelically expressed and in mice, about 3%. Expression from the inactive X allele varies from a few percent of that from the active allele to near equal expression. While most genes have a stable inactivation pattern, a subset of genes exhibit tissue-specific differences in escape from X inactivation. Escape genes appear to be protected from the repressive chromatin modifications associated with X inactivation. Differences in the identity and distribution of escape genes between species and tissues suggest a role for these genes in the evolution of sex differences in specific phenotypes. The higher expression of escape genes in females than in males implies that they may have female-specific roles and may be responsible for some of the phenotypes observed in X aneuploidy.
Conflict of interest statement
Figures

References
-
- Abidi F, Holloway L, Moore CA, Weaver DD, Simensen RJ, Stevenson RE, Rogers RC, et al. Novel human pathological mutations. Gene symbol: JARID1C. Disease: mental retardation, X-linked. Hum Genet. 2009;125(3):345. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

