Cellular dynamics of RNA modification
- PMID: 21615108
- PMCID: PMC3179539
- DOI: 10.1021/ar200057m
Cellular dynamics of RNA modification
Abstract
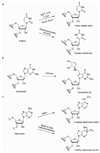
Five decades of research have identified more than 100 ribonucleosides that are post-transcriptionally modified. Many modified nucleosides are conserved throughout bacteria, archaea, and eukaryotes, while some are unique to each branch of life. However, the cellular and functional dynamics of RNA modification remain largely unexplored, mostly because of the lack of functional hypotheses and experimental methods for quantification and large-scale analysis. Many RNA modifications are not essential for life, which parallels the observation that many well-characterized protein and DNA modifications are not essential for life. Instead, increasing evidence indicates that RNA modifications can play regulatory roles in cells, especially in response to stress conditions. In this Account, we review some examples of RNA modification that are dynamically controlled in cells. We also discuss some recently developed methods that have enhanced the ability to study the cellular dynamics of RNA modification. We discuss four specific examples of RNA modification in detail here. We begin with 4-thio uridine (s(4)U), which can act as a cellular sensor of near-UV light. Then we consider queuosine (Q), which is a potential biomarker for malignancy. Next we examine N(6)-methyl adenine (m(6)A), which is the prevalent modification in eukaryotic messenger RNAs (mRNAs). Finally, we discuss pseudouridine (ψ), which is inducible by nutrient deprivation. We then consider two recent technical advances that have stimulated the study of the cellular dynamics in modified ribonucleosides. The first is a genome-wide method that combines primer extension with a microarray. It was used to study the N(1)-methyl adenine (m(1)A) hypomodification in human transfer RNA (tRNA). The second is a quantitative mass spectrometric method used to investigate dynamic changes in a wide range of tRNA modifications under stress conditions in yeast. In addition, we discuss potential mechanisms that control dynamic regulation of RNA modifications as well as hypotheses for discovering potential RNA demodification enzymes. We conclude by highlighting the need to develop new tools and to generate additional hypotheses for how these modifications function in cells. The study of the cellular dynamics of modified RNA remains a largely open area for new development, which underscores the rich potential for important advances as researchers drive this emerging field to the next level.
Figures



References
-
- Grosjean H. Modification and editing of RNA: historical overview and important facts to remember. In: Grosjean H, editor. Fine-tuning of RNA functions by modification and editing. Berlin: Springer Verlag; 2005. pp. 1–22.
-
- Grosjean H, Benne R, editors. Modification and Editing of RNA. Washington, DC: ASM press; 1998.
-
- Grosjean H, editor. Fine-tuning of RNA functions by modification and editing. Berlin: Springer-Verlag; 2005.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

