Progress in structure based drug design for G protein-coupled receptors
- PMID: 21615150
- PMCID: PMC3308205
- DOI: 10.1021/jm200371q
Progress in structure based drug design for G protein-coupled receptors
Figures
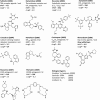
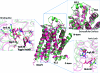

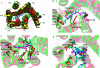



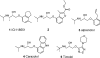
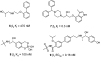
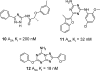
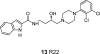

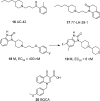
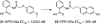

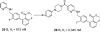


References
-
- Bikker J. A.; Trumpp-Kallmeyer S.; Humblet C. G-Protein coupled receptors: models, mutagenesis, and drug design. J. Med. Chem. 1998, 41 (16), 2911–2927. - PubMed
-
- Henderson R.; Schertler G. F. The structure of bacteriorhodopsin and its relevance to the visual opsins and other seven-helix G-protein coupled receptors. Philos. Trans. R. Soc. London, Ser. B 1990, 326 (1236), 379–389. - PubMed
-
- Schertler G. F.; Villa C.; Henderson R. Projection structure of rhodopsin. Nature 1993, 362 (6422), 770–772. - PubMed
-
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 2001, 46 (1–3), 3–26. - PubMed
-
- Leeson P. D.; Springthorpe B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat. Rev. Drug Discovery 2007, 6 (11), 881–890. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

