P53 in human melanoma fails to regulate target genes associated with apoptosis and the cell cycle and may contribute to proliferation
- PMID: 21615965
- PMCID: PMC3120805
- DOI: 10.1186/1471-2407-11-203
P53 in human melanoma fails to regulate target genes associated with apoptosis and the cell cycle and may contribute to proliferation
Abstract
Background: Metastatic melanoma represents a major clinical problem. Its incidence continues to rise in western countries and there are currently no curative treatments. While mutation of the P53 tumour suppressor gene is a common feature of many types of cancer, mutational inactivation of P53 in melanoma is uncommon; however, its function often appears abnormal.
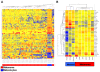
Methods: In this study whole genome bead arrays were used to examine the transcript expression of P53 target genes in extracts from 82 melanoma metastases and 6 melanoma cell lines, to provide a global assessment of aberrant P53 function. The expression of these genes was also examined in extracts derived from diploid human melanocytes and fibroblasts.
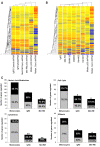
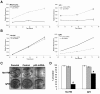
Results: The results indicated that P53 target transcripts involved in apoptosis were under-expressed in melanoma metastases and melanoma cell lines, while those involved in the cell cycle were over-expressed in melanoma cell lines. There was little difference in the transcript expression of P53 target genes between cell lines with null/mutant P53 compared to those with wild-type P53, suggesting that altered expression in melanoma was not related to P53 status. Similarly, down-regulation of P53 by short-hairpin RNA (shRNA) had limited effect on P53 target gene expression in melanoma cells, whereas there were a large number of P53 target genes whose mRNA expression was significantly altered by P53 inhibition in melanocytes. Analysis of whole genome gene expression profiles indicated that the ability of P53 to regulate genes involved in the cell cycle was significantly reduced in melanoma cells. Moreover, inhibition of P53 in melanocytes induced changes in gene expression profiles that were characteristic of melanoma cells and resulted in increased proliferation. Conversely, knockdown of P53 in melanoma cells resulted in decreased proliferation.
Conclusions: These results indicate that P53 target genes involved in apoptosis and cell cycle regulation are aberrantly expressed in melanoma and that this aberrant functional activity of P53 may contribute to the proliferation of melanoma.
Figures

References
-
- Thompson JF, Scolyer RA, Kefford RF. Cutaneous melanoma. Lancet. 2005;365:687–701. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

