DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase
- PMID: 21620135
- PMCID: PMC3129610
- DOI: 10.1016/j.cell.2011.04.015
DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase
Abstract
G-quadruplex (G4) DNA structures are extremely stable four-stranded secondary structures held together by noncanonical G-G base pairs. Genome-wide chromatin immunoprecipitation was used to determine the in vivo binding sites of the multifunctional Saccharomyces cerevisiae Pif1 DNA helicase, a potent unwinder of G4 structures in vitro. G4 motifs were a significant subset of the high-confidence Pif1-binding sites. Replication slowed in the vicinity of these motifs, and they were prone to breakage in Pif1-deficient cells, whereas non-G4 Pif1-binding sites did not show this behavior. Introducing many copies of G4 motifs caused slow growth in replication-stressed Pif1-deficient cells, which was relieved by spontaneous mutations that eliminated their ability to form G4 structures, bind Pif1, slow DNA replication, and stimulate DNA breakage. These data suggest that G4 structures form in vivo and that they are resolved by Pif1 to prevent replication fork stalling and DNA breakage.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


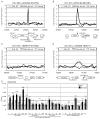



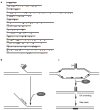
Comment in
-
DNA replication: Pif1 overcomes a quadruplex hurdle.Nat Rev Mol Cell Biol. 2011 Jun 15;12(7):402. doi: 10.1038/nrm3142. Nat Rev Mol Cell Biol. 2011. PMID: 21673725 No abstract available.
References
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JRoy Statist Soc Ser B. 1995;57:289–300.
-
- Boule J, Vega L, Zakian V. The Yeast Pif1p helicase removes telomerase from DNA. Nature. 2005;438:57–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

