Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1
- PMID: 21620138
- PMCID: PMC3130564
- DOI: 10.1016/j.cell.2011.03.054
Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1
Abstract
The pyruvate kinase isoforms PKM1 and PKM2 are alternatively spliced products of the PKM2 gene. PKM2, but not PKM1, alters glucose metabolism in cancer cells and contributes to tumorigenesis by mechanisms that are not explained by its known biochemical activity. We show that PKM2 gene transcription is activated by hypoxia-inducible factor 1 (HIF-1). PKM2 interacts directly with the HIF-1α subunit and promotes transactivation of HIF-1 target genes by enhancing HIF-1 binding and p300 recruitment to hypoxia response elements, whereas PKM1 fails to regulate HIF-1 activity. Interaction of PKM2 with prolyl hydroxylase 3 (PHD3) enhances PKM2 binding to HIF-1α and PKM2 coactivator function. Mass spectrometry and anti-hydroxyproline antibody assays demonstrate PKM2 hydroxylation on proline-403/408. PHD3 knockdown inhibits PKM2 coactivator function, reduces glucose uptake and lactate production, and increases O(2) consumption in cancer cells. Thus, PKM2 participates in a positive feedback loop that promotes HIF-1 transactivation and reprograms glucose metabolism in cancer cells.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


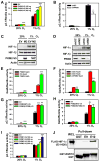

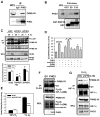
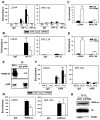
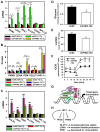
Comment in
-
PK-M2 Makes Cells Sweeter on HIF1.Cell. 2011 May 27;145(5):647-9. doi: 10.1016/j.cell.2011.05.009. Cell. 2011. PMID: 21620132
-
Cancer metabolism: feed it forward.Nat Rev Cancer. 2011 Jun 9;11(7):461. doi: 10.1038/nrc3094. Nat Rev Cancer. 2011. PMID: 21654817 No abstract available.
References
-
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. - PubMed
-
- Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

