Dimer interface of the effector domain of non-structural protein 1 from influenza A virus: an interface with multiple functions
- PMID: 21622573
- PMCID: PMC3138300
- DOI: 10.1074/jbc.M111.248765
Dimer interface of the effector domain of non-structural protein 1 from influenza A virus: an interface with multiple functions
Abstract
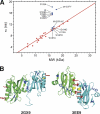
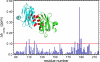
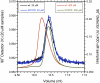
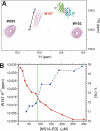
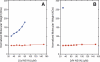
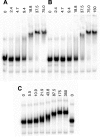
Non-structural protein 1 from influenza A virus, NS1A, is a key multifunctional virulence factor composed of two domains: an N-terminal double-stranded RNA (dsRNA)-binding domain and a C-terminal effector domain (ED). Isolated RNA-binding and effector domains of NS1A both exist as homodimers in solution. Despite recent crystal structures of isolated ED and full-length NS1A proteins from different influenza virus strains, controversy remains over the actual biologically relevant ED dimer interface. Here, we report the biophysical properties of the NS1A ED from H3N2 influenza A/Udorn/307/1972 (Ud) virus in solution. Several lines of evidence, including (15)N NMR relaxation, NMR chemical shift perturbations, static light scattering, and analytical sedimentation equilibrium, demonstrate that Ud NS1A ED forms a relatively weak dimer in solution (K(d) = 90 ± 2 μm), featuring a symmetric helix-helix dimer interface. Mutations within and near this interface completely abolish dimerization, whereas mutations consistent with other proposed ED dimer interfaces have no effect on dimer formation. In addition, the critical Trp-187 residue in this interface serves as a sensitive NMR spectroscopic marker for the concentration-dependent dimerization of NS1A ED in solution. Finally, dynamic light scattering and gel shift binding experiments demonstrate that the ED interface plays a role in both the oligomerization and the dsRNA binding properties of the full-length NS1A protein. In particular, mutation of the critical tryptophan in the ED interface substantially reduces the propensity of full-length NS1A from different strains to oligomerize and results in a reduction in dsRNA binding affinity for full-length NS1A.
Figures



References
-
- Hale B. G., Randall R. E., Ortín J., Jackson D. (2008) J. Gen. Virol. 89, 2359–2376 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

