Cholesterol degradation by Gordonia cholesterolivorans
- PMID: 21622796
- PMCID: PMC3147395
- DOI: 10.1128/AEM.05149-11
Cholesterol degradation by Gordonia cholesterolivorans
Abstract
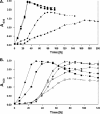
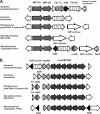
This paper reports physiological and genetic data about the type strain Gordonia cholesterolivorans, a strain that is able to degrade steroid compounds containing a long carbon side chain such as cholesterol (C(27)), cholestenone (C(27)), ergosterol (C(28)), and stigmasterol (C(29)). The length of the carbon side chain appears to be of great importance for this bacterium, as the strain is unable to grow using steroids with a shorter or nonaliphatic carbon side chain such as cholic acid (C(24)), progesterone (C(21)), testosterone, androsterone, 4-androstene-3,17-dione (all C(19)), and further steroids. This study also demonstrates that the degradation of cholesterol is a quite common feature of the genus Gordonia by comparing Gordonia cholesterolivorans with some other species of this genus (e.g., G. sihwensis, G. hydrophobica, G. australis, and G. neofelifaecis). Pyrosequencing of the genome of G. cholesterolivorans led to the identification of two conventional cholesterol oxidase genes on an 8-kb and a 12.8-kb genomic fragment with genetic organizations that are quite unique as compared to the genomes of other cholesterol-degrading bacteria sequenced so far. The identified two putative cholesterol oxidases of G. cholesterolivorans are both intracellularly acting enzymes of the class I type. Whereas one of these two cholesterol oxidases (ChoOx-1) shows high identity with an oxidoreductase of the opportunistic pathogen G. bronchialis and is not transcribed during growth with cholesterol, the other one (ChoOx-2) appears phylogenetically closer to cholesterol oxidases from members of the genus Rhodococcus and is transcribed constitutively. By using targeted gene disruption, a G. cholesterolivorans ChoOx-2 gene mutant strain that was unable to grow with steroids was obtained.
Figures




References
-
- Blaschke A. J., et al. 2007. Gordonia species: emerging pathogens in pediatric patients that are identified by 16S ribosomal RNA gene sequencing. Clin. Infect. Dis. 45:483–486 - PubMed
-
- Cavener D. R. 1992. GMC oxidoreductases. A newly defined family of homologous proteins with diverse catalytic activities. J. Mol. Biol. 223:811–814 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

