Impact of exercise training on endothelial transcriptional profiles in healthy swine: a genome-wide microarray analysis
- PMID: 21622830
- PMCID: PMC3154664
- DOI: 10.1152/ajpheart.00065.2011
Impact of exercise training on endothelial transcriptional profiles in healthy swine: a genome-wide microarray analysis
Abstract
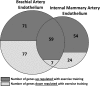
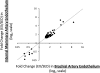
While the salutary effects of exercise training on conduit artery endothelial cells have been reported in animals and humans with cardiovascular risk factors or disease, whether a healthy endothelium is alterable with exercise training is less certain. The purpose of this study was to evaluate the impact of exercise training on transcriptional profiles in normal endothelial cells using a genome-wide microarray analysis. Brachial and internal mammary endothelial gene expression was compared between a group of healthy pigs that exercise trained for 16-20 wk (n = 8) and a group that remained sedentary (n = 8). We found that a total of 130 genes were upregulated and 84 genes downregulated in brachial artery endothelial cells with exercise training (>1.5-fold and false discovery rate <15%). In contrast, a total of 113 genes were upregulated and 31 genes downregulated in internal mammary artery endothelial cells using the same criteria. Although there was an overlap of 66 genes (59 upregulated and 7 downregulated with exercise training) between the brachial and internal mammary arteries, the identified endothelial gene networks and biological processes influenced by exercise training were distinctly different between the brachial and internal mammary arteries. These data indicate that a healthy endothelium is indeed responsive to exercise training and support the concept that the influence of physical activity on endothelial gene expression is not homogenously distributed throughout the vasculature.
Figures



References
-
- Aird WC. Endothelial cell heterogeneity and atherosclerosis. Curr Atheroscler Rep 8: 69–75, 2006 - PubMed
-
- Aird WC. Spatial and temporal dynamics of the endothelium. J Thromb Haemost 3: 1392–1406, 2005 - PubMed
-
- Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607, 2006 - PubMed
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29, 2000 - PMC - PubMed
-
- Barone R. (Editor). Anatomie Comparee des Mammiferes Domestiques. Paris: Vigot; 1996
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

