The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains
- PMID: 21623345
- PMCID: PMC3155300
- DOI: 10.1038/emboj.2011.166
The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains
Abstract
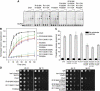
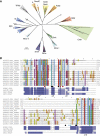
The ATP-dependent chromatin-remodelling enzyme Chd1 is a 168-kDa protein consisting of a double chromodomain, Snf2-related ATPase domain, and a C-terminal DNA-binding domain. Here, we show the DNA-binding domain is required for Saccharomyces cerevisiae Chd1 to bind and remodel nucleosomes. The crystal structure of this domain reveals the presence of structural homology to SANT and SLIDE domains previously identified in ISWI remodelling enzymes. The presence of these domains in ISWI and Chd1 chromatin-remodelling enzymes may provide a means of efficiently harnessing the action of the Snf2-related ATPase domain for the purpose of nucleosome spacing and provide an explanation for partial redundancy between these proteins. Site directed mutagenesis was used to identify residues important for DNA binding and generate a model describing the interaction of this domain with DNA. Through inclusion of Chd1 sequences in homology searches SLIDE domains were identified in CHD6-9 proteins. Point mutations to conserved amino acids within the human CHD7 SLIDE domain have been identified in patients with CHARGE syndrome.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Aasland R, Stewart AF, Gibson T (1996) The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci 21: 87–88 - PubMed
-
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 - PMC - PubMed
-
- Alen C, Kent NA, Jones HS, O’Sullivan J, Aranda A, Proudfoot NJ (2002) A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol Cell 10: 1441–1452 - PubMed
-
- Bailey S (1994) The Ccp4 suite—programs for protein crystallography. Acta Crystallogr D 50: 760–763 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

