Latent TGF-β binding protein 3 identifies a second heart field in zebrafish
- PMID: 21623370
- PMCID: PMC3319150
- DOI: 10.1038/nature10094
Latent TGF-β binding protein 3 identifies a second heart field in zebrafish
Abstract
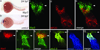
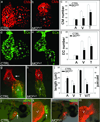
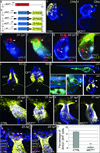
The four-chambered mammalian heart develops from two fields of cardiac progenitor cells distinguished by their spatiotemporal patterns of differentiation and contributions to the definitive heart. The first heart field differentiates earlier in lateral plate mesoderm, generates the linear heart tube and ultimately gives rise to the left ventricle. The second heart field (SHF) differentiates later in pharyngeal mesoderm, elongates the heart tube, and gives rise to the outflow tract and much of the right ventricle. Because hearts in lower vertebrates contain a rudimentary outflow tract but not a right ventricle, the existence and function of SHF-like cells in these species has remained a topic of speculation. Here we provide direct evidence from Cre/Lox-mediated lineage tracing and loss-of-function studies in zebrafish, a lower vertebrate with a single ventricle, that latent TGF-β binding protein 3 (ltbp3) transcripts mark a field of cardiac progenitor cells with defining characteristics of the anterior SHF in mammals. Specifically, ltbp3(+) cells differentiate in pharyngeal mesoderm after formation of the heart tube, elongate the heart tube at the outflow pole, and give rise to three cardiovascular lineages in the outflow tract and myocardium in the distal ventricle. In addition to expressing Ltbp3, a protein that regulates the bioavailability of TGF-β ligands, zebrafish SHF cells co-express nkx2.5, an evolutionarily conserved marker of cardiac progenitor cells in both fields. Embryos devoid of ltbp3 lack the same cardiac structures derived from ltbp3(+) cells due to compromised progenitor proliferation. Furthermore, small-molecule inhibition of TGF-β signalling phenocopies the ltbp3-morphant phenotype whereas expression of a constitutively active TGF-β type I receptor rescues it. Taken together, our findings uncover a requirement for ltbp3-TGF-β signalling during zebrafish SHF development, a process that serves to enlarge the single ventricular chamber in this species.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

