Multi-walled carbon nanotube-induced gene expression in the mouse lung: association with lung pathology
- PMID: 21624382
- PMCID: PMC3148292
- DOI: 10.1016/j.taap.2011.05.012
Multi-walled carbon nanotube-induced gene expression in the mouse lung: association with lung pathology
Abstract
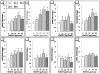
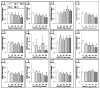
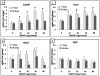
Due to the fibrous shape and durability of multi-walled carbon nanotubes (MWCNT), concerns regarding their potential for producing environmental and human health risks, including carcinogenesis, have been raised. This study sought to investigate how previously identified lung cancer prognostic biomarkers and the related cancer signaling pathways are affected in the mouse lung following pharyngeal aspiration of well-dispersed MWCNT. A total of 63 identified lung cancer prognostic biomarker genes and major signaling biomarker genes were analyzed in mouse lungs (n=80) exposed to 0, 10, 20, 40, or 80μg of MWCNT by pharyngeal aspiration at 7 and 56days post-exposure using quantitative PCR assays. At 7 and 56days post-exposure, a set of 7 genes and a set of 11 genes, respectively, showed differential expression in the lungs of mice exposed to MWCNT vs. the control group. Additionally, these significant genes could separate the control group from the treated group over the time series in a hierarchical gene clustering analysis. Furthermore, 4 genes from these two sets of significant genes, coiled-coil domain containing-99 (Ccdc99), muscle segment homeobox gene-2 (Msx2), nitric oxide synthase-2 (Nos2), and wingless-type inhibitory factor-1 (Wif1), showed significant mRNA expression perturbations at both time points. It was also found that the expression changes of these 4 overlapping genes at 7days post-exposure were attenuated at 56days post-exposure. Ingenuity Pathway Analysis (IPA) found that several carcinogenic-related signaling pathways and carcinogenesis itself were associated with both the 7 and 11 gene signatures. Taken together, this study identifies that MWCNT exposure affects a subset of lung cancer biomarkers in mouse lungs.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures








References
-
- Aubrecht J, Caba E. Gene expression profile analysis: an emerging approach to investigate mechanisms of genotoxicity. Pharmacogenomics. 2005;6:419–428. - PubMed
-
- Auerbach O, Garfinkel L, Parks VR. Scar cancer of the lung: increase over a 21 year period. Cancer. 1979;43:636–642. - PubMed
-
- Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

