EBP1 and DRBP76/NF90 binding proteins are included in the major histocompatibility complex class II RNA operon
- PMID: 21624892
- PMCID: PMC3167597
- DOI: 10.1093/nar/gkr278
EBP1 and DRBP76/NF90 binding proteins are included in the major histocompatibility complex class II RNA operon
Abstract
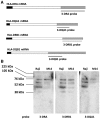
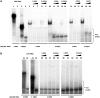
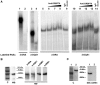
Major histocompatibility complex class II mRNAs encode heterodimeric proteins involved in the presentation of exogenous antigens during an immune response. Their 3'UTRs bind a protein complex in which we identified two factors: EBP1, an ErbB3 receptor-binding protein and DRBP76, a double-stranded RNA binding nuclear protein, also known as nuclear factor 90 (NF90). Both are well-characterized regulatory factors of several mRNA molecules processing. Using either EBP1 or DRBP76/NF90-specific knockdown experiments, we established that the two proteins play a role in regulating the expression of HLA-DRA, HLA-DRB1 and HLA-DQA1 mRNAs levels. Our study represents the first indication of the existence of a functional unit that includes different transcripts involved in the adaptive immune response. We propose that the concept of 'RNA operon' may be suitable for our system in which MHCII mRNAs are modulated via interaction of their 3'UTR with same proteins.
Figures


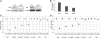

References
-
- Cresswell P. Assembly, transport, and function of MHC class II molecules. Annu. Rev. Immunol. 1994;12:259–293. - PubMed
-
- Rodriguez T, Mendez R, Del Campo A, Aptsiauri N, Martin J, Orozco G, Pawelec G, Schadendorf D, Ruiz-Cabello F, Garrido F. Patterns of constitutive and IFN-gamma inducible expression of HLA class II molecules in human melanoma cell lines. Immunogenetics. 2007;59:123–133. - PubMed
-
- Guardiola J, Maffei A. Control of MHC class II gene expression in autoimmune, infectious, and neoplastic diseases. Crit. Rev. Immunol. 1993;13:247–268. - PubMed
-
- Reith W, LeibundGut-Landmann S, Waldburger JM. Regulation of MHC class II gene expression by the class II transactivator. Nat. Rev. Immunol. 2005;5:793–806. - PubMed
-
- Tite JP, Janeway CA., Jr Antigen-dependent selection of B lymphoma cells varying in Ia density by cloned antigen-specific L3T4a+ T cells: a possible in vitro model for B cell adaptive differentiation. J. Mol. Cell Immunol. 1984;1:253–265. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

