RAG: an update to the RNA-As-Graphs resource
- PMID: 21627789
- PMCID: PMC3123240
- DOI: 10.1186/1471-2105-12-219
RAG: an update to the RNA-As-Graphs resource
Abstract
Background: In 2004, we presented a web resource for stimulating the search for novel RNAs, RNA-As-Graphs (RAG), which classified, catalogued, and predicted RNA secondary structure motifs using clustering and build-up approaches. With the increased availability of secondary structures in recent years, we update the RAG resource and provide various improvements for analyzing RNA structures.
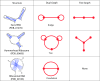
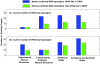
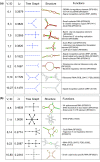
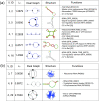
Description: Our RAG update includes a new supervised clustering algorithm that can suggest RNA motifs that may be "RNA-like". We use this utility to describe RNA motifs as three classes: existing, RNA-like, and non-RNA-like. This produces 126 tree and 16,658 dual graphs as candidate RNA-like topologies using the supervised clustering algorithm with existing RNAs serving as the training data. A comparison of this clustering approach to an earlier method shows considerable improvements. Additional RAG features include greatly expanded search capabilities, an interface to better utilize the benefits of relational database, and improvements to several of the utilities such as directed/labeled graphs and a subgraph search program.
Conclusions: The RAG updates presented here augment the database's intended function - stimulating the search for novel RNA functionality - by classifying available motifs, suggesting new motifs for design, and allowing for more specific searches for specific topologies. The updated RAG web resource offers users a graph-based tool for exploring available RNA motifs and suggesting new RNAs for design.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

