The development and characterization of a 60K SNP chip for chicken
- PMID: 21627800
- PMCID: PMC3117858
- DOI: 10.1186/1471-2164-12-274
The development and characterization of a 60K SNP chip for chicken
Abstract
Background: In livestock species like the chicken, high throughput single nucleotide polymorphism (SNP) genotyping assays are increasingly being used for whole genome association studies and as a tool in breeding (referred to as genomic selection). To be of value in a wide variety of breeds and populations, the success rate of the SNP genotyping assay, the distribution of the SNP across the genome and the minor allele frequencies (MAF) of the SNPs used are extremely important.
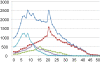
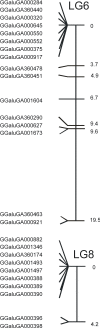
Results: We describe the design of a moderate density (60k) Illumina SNP BeadChip in chicken consisting of SNPs known to be segregating at high to medium minor allele frequencies (MAF) in the two major types of commercial chicken (broilers and layers). This was achieved by the identification of 352,303 SNPs with moderate to high MAF in 2 broilers and 2 layer lines using Illumina sequencing on reduced representation libraries. To further increase the utility of the chip, we also identified SNPs on sequences currently not covered by the chicken genome assembly (Gallus_gallus-2.1). This was achieved by 454 sequencing of the chicken genome at a depth of 12x and the identification of SNPs on 454-derived contigs not covered by the current chicken genome assembly. In total we added 790 SNPs that mapped to 454-derived contigs as well as 421 SNPs with a position on Chr_random of the current assembly. The SNP chip contains 57,636 SNPs of which 54,293 could be genotyped and were shown to be segregating in chicken populations. Our SNP identification procedure appeared to be highly reliable and the overall validation rate of the SNPs on the chip was 94%. We were able to map 328 SNPs derived from the 454 sequence contigs on the chicken genome. The majority of these SNPs map to chromosomes that are already represented in genome build Gallus_gallus-2.1.0. Twenty-eight SNPs were used to construct two new linkage groups most likely representing two micro-chromosomes not covered by the current genome assembly.
Conclusions: The high success rate of the SNPs on the Illumina chicken 60K Beadchip emphasizes the power of Next generation sequence (NGS) technology for the SNP identification and selection step. The identification of SNPs from sequence contigs derived from NGS sequencing resulted in improved coverage of the chicken genome and the construction of two new linkage groups most likely representing two chicken micro-chromosomes.
Figures

References
-
- Magee DA, Park SD, Scraggs E, Murphy AM, Doherty ML, Kijas JW. International Sheep Genomics Consortium. MacHugh DE. Technical note: High fidelity of whole-genome amplified sheep (Ovis aries) deoxyribonucleic acid using a high-density single nucleotide polymorphism array-based genotyping platform. J Anim Sci. 2010;88:3183–3186. doi: 10.2527/jas.2009-2723. - DOI - PubMed
-
- Ramos AM, Crooijmans RPMA, Affara NA, Amaral AJ, Archibald AL, Beever JE, Bendixen C, Churcher C, Clark R, Dehais P, Hansen MS, Hedegaard J, Hu Z-L, Kerstens HH, Law AS, Megens H-J, Milan D, Nonneman DJ, Rohrer GA, Rothschild MF, Smith TPL, Schnabel RD, Van Tassell CP, Taylor JF, Wiedmann RT, Schook LB, Groenen MAM. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One. 2009;4:e6524. doi: 10.1371/journal.pone.0006524. - DOI - PMC - PubMed
-
- Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MAM, Delany ME, Dodgson JB, Chinwalla AT, Cliften PF, Clifton SW, Delehaunty KD, Fronick C, Fulton RS, Graves TA, Kremitzki C, Layman D, Magrini V, McPherson JD, Miner TL, Minx P, Nash WE, Nhan MN, Nelson JO, Oddy LG, Pohl CS, Randall-Maher J. International Chicken Genome Sequencing Consortium et al.Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. - DOI - PubMed
-
- Wong GK, Liu B, Wang J, Zhang Y, Yang X, Zhang Z, Meng Q, Zhou J, Li D, Zhang J, Ni P, Li S, Ran L, Li H, Zhang J, Li R, Li S, Zheng H, Lin W, Li G, Wang X, Zhao W, Li J, Ye C, Dai M, Ruan J, Zhou Y, Li Y, He X, Zhang Y. International Chicken Polymorphism Map Consortium et al.A genetic variation map for chicken with 28 million single-nucleotide polymorphisms. Nature. 2004;432:717–722. doi: 10.1038/nature03156. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

