Song exposure regulates known and novel microRNAs in the zebra finch auditory forebrain
- PMID: 21627805
- PMCID: PMC3118218
- DOI: 10.1186/1471-2164-12-277
Song exposure regulates known and novel microRNAs in the zebra finch auditory forebrain
Abstract
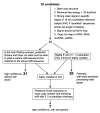
Background: In an important model for neuroscience, songbirds learn to discriminate songs they hear during tape-recorded playbacks, as demonstrated by song-specific habituation of both behavioral and neurogenomic responses in the auditory forebrain. We hypothesized that microRNAs (miRNAs or miRs) may participate in the changing pattern of gene expression induced by song exposure. To test this, we used massively parallel Illumina sequencing to analyse small RNAs from auditory forebrain of adult zebra finches exposed to tape-recorded birdsong or silence.
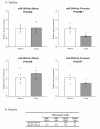
Results: In the auditory forebrain, we identified 121 known miRNAs conserved in other vertebrates. We also identified 34 novel miRNAs that do not align to human or chicken genomes. Five conserved miRNAs showed significant and consistent changes in copy number after song exposure across three biological replications of the song-silence comparison, with two increasing (tgu-miR-25, tgu-miR-192) and three decreasing (tgu-miR-92, tgu-miR-124, tgu-miR-129-5p). We also detected a locus on the Z sex chromosome that produces three different novel miRNAs, with supporting evidence from Northern blot and TaqMan qPCR assays for differential expression in males and females and in response to song playbacks. One of these, tgu-miR-2954-3p, is predicted (by TargetScan) to regulate eight song-responsive mRNAs that all have functions in cellular proliferation and neuronal differentiation.
Conclusions: The experience of hearing another bird singing alters the profile of miRNAs in the auditory forebrain of zebra finches. The response involves both known conserved miRNAs and novel miRNAs described so far only in the zebra finch, including a novel sex-linked, song-responsive miRNA. These results indicate that miRNAs are likely to contribute to the unique behavioural biology of learned song communication in songbirds.
Figures

References
-
- Miller DB. Acoustic Basis of Mate Recognition by Female Zebra Finches (Taeniopygia-Guttata) Animal Behaviour. 1979;27(May):376–380.
-
- Miller DB. Long-Term Recognition of Fathers Song by Female Zebra Finches. Nature. 1979;280(5721):389–391. doi: 10.1038/280389a0. - DOI
-
- Clayton NS. Song Discrimination-Learning in Zebra Finches. Animal Behaviour. 1988;36:1016–1024. doi: 10.1016/S0003-3472(88)80061-7. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

