Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with Total Therapy 3
- PMID: 21628408
- PMCID: PMC3186329
- DOI: 10.1182/blood-2010-12-328252
Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with Total Therapy 3
Abstract
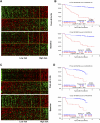
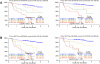
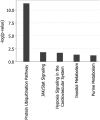
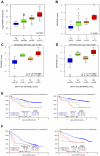
Gene expression profiling (GEP) of purified plasma cells 48 hours after thalidomide and dexamethasone test doses showed these agents' mechanisms of action and provided prognostic information for untreated myeloma patients on Total Therapy 2 (TT2). Bortezomib was added in Total Therapy 3 (TT3), and 48 hours after bortezomib GEP analysis identified 80 highly survival-discriminatory genes in a training set of 142 TT3A patients that were validated in 128 patients receiving TT3B. The 80-gene GEP model (GEP80) also distinguished outcomes when applied at baseline in both TT3 and TT2 protocols. In context of our validated 70-gene model (GEP70), the GEP80 model identified 9% of patients with a grave prognosis among those with GEP70-defined low-risk disease and 41% of patients with favorable prognosis among those with GEP70-defined high-risk disease. PMSD4 was 1 of 3 genes common to both models. Residing on chromosome 1q21, PSMD4 expression is highly sensitive to copy number. Both higher PSMD4 expression levels and higher 1q21 copy numbers affected clinical outcome adversely. GEP80 baseline-defined high risk, high lactate dehydrogenase, and low albumin were the only independent adverse variables surviving multivariate survival model. We are investigating whether second-generation proteasome inhibitors (eg, carfilzomib) can overcome resistance associated with high PSMD4 levels.
Figures



References
-
- Palumbo A, Bringhen S, Liberati AM, et al. Oral melphalan, prednisone, and thalidomide in elderly patients with multiple myeloma: updated results of a randomized controlled trial. Blood. 2008;112(8):3107–3114. - PubMed
-
- Palumbo A, Falco P, Corradini P, et al. Melphalan, prednisone, and lenalidomide treatment for newly diagnosed myeloma: a report from the GIMEMA–Italian Multiple Myeloma Network. J Clin Oncol. 2007;25(28):4459–4465. - PubMed
-
- San Miguel JF, Schlag R, Khuageva NK, et al. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N Engl J Med. 2008;359(9):906–917. - PubMed
-
- Mateos MV, Oriol A, Martinez-Lopez J, et al. Bortezomib, melphalan, and prednisone versus bortezomib, thalidomide, and prednisone as induction therapy followed by maintenance treatment with bortezomib and thalidomide versus bortezomib and prednisone in elderly patients with untreated multiple myeloma: a randomised trial. Lancet Oncol. 2010;11(10):934–941. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

