Cell type-specific DNA methylation at intragenic CpG islands in the immune system
- PMID: 21628449
- PMCID: PMC3129250
- DOI: 10.1101/gr.118703.110
Cell type-specific DNA methylation at intragenic CpG islands in the immune system
Abstract
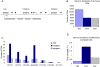
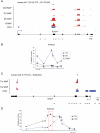
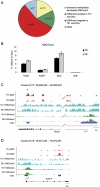
Human and mouse genomes contain a similar number of CpG islands (CGIs), which are discrete CpG-rich DNA sequences associated with transcription start sites. In both species, ∼50% of all CGIs are remote from annotated promoters but, nevertheless, often have promoter-like features. To determine the role of CGI methylation in cell differentiation, we analyzed DNA methylation at a comprehensive CGI set in cells of the mouse hematopoietic lineage. Using a method that potentially detects ∼33% of genomic CpGs in the methylated state, we found that large differences in gene expression were accompanied by surprisingly few DNA methylation changes. There were, however, many DNA methylation differences between hematopoietic cells and a distantly related tissue, brain. Altered DNA methylation in the immune system occurred predominantly at CGIs within gene bodies, which have the properties of cell type-restricted promoters, but infrequently at annotated gene promoters or CGI flanking sequences (CGI "shores"). Unexpectedly, elevated intragenic CGI methylation correlated with silencing of the associated gene. Differentially methylated intragenic CGIs tended to lack H3K4me3 and associate with a transcriptionally repressive environment regardless of methylation state. Our results indicate that DNA methylation changes play a relatively minor role in the late stages of differentiation and suggest that intragenic CGIs represent regulatory sites of differential gene expression during the early stages of lineage specification.
Figures



References
-
- Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. 2009. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol 16: 717–724 - PubMed
-
- Antequera F, Boyes J, Bird A 1990. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell 62: 503–514 - PubMed
-
- Batsche E, Yaniv M, Muchardt C 2006. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol 13: 22–29 - PubMed
-
- Begenisich T, Nakamoto T, Ovitt CE, Nehrke K, Brugnara C, Alper SL, Melvin JE 2004. Physiological roles of the intermediate conductance, Ca2+-activated potassium channel Kcnn4. J Biol Chem 279: 47681–47687 - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
