Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor
- PMID: 21631110
- PMCID: PMC3126897
- DOI: 10.1021/ja202902d
Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor
Abstract
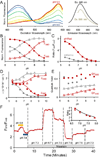
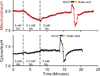
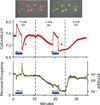
Intracellular pH affects protein structure and function, and proton gradients underlie the function of organelles such as lysosomes and mitochondria. We engineered a genetically encoded pH sensor by mutagenesis of the red fluorescent protein mKeima, providing a new tool to image intracellular pH in live cells. This sensor, named pHRed, is the first ratiometric, single-protein red fluorescent sensor of pH. Fluorescence emission of pHRed peaks at 610 nm while exhibiting dual excitation peaks at 440 and 585 nm that can be used for ratiometric imaging. The intensity ratio responds with an apparent pK(a) of 6.6 and a >10-fold dynamic range. Furthermore, pHRed has a pH-responsive fluorescence lifetime that changes by ~0.4 ns over physiological pH values and can be monitored with single-wavelength two-photon excitation. After characterizing the sensor, we tested pHRed's ability to monitor intracellular pH by imaging energy-dependent changes in cytosolic and mitochondrial pH.
Figures

References
-
- Casey JR, Grinstein S, Orlowski J. Nat. Rev. Mol. Cell Biol. 2010;11:50–61. - PubMed
-
- Lagadic-Gossmann D, Huc L, Lecureur V. Cell Death Differ. 2004;11:953–961. - PubMed
-
- Han J, Burgess K. Chem. Rev. 2010;110:2709–2928. - PubMed
-
- Bizzarri R, Serresi M, Luin S, Beltram F. Anal. Bioanal. Chem. 2009;393:1107–1122. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

