A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas
- PMID: 21632744
- PMCID: PMC3186192
- DOI: 10.1074/mcp.M110.006353
A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas
Abstract
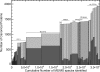
Human blood plasma can be obtained relatively noninvasively and contains proteins from most, if not all, tissues of the body. Therefore, an extensive, quantitative catalog of plasma proteins is an important starting point for the discovery of disease biomarkers. In 2005, we showed that different proteomics measurements using different sample preparation and analysis techniques identify significantly different sets of proteins, and that a comprehensive plasma proteome can be compiled only by combining data from many different experiments. Applying advanced computational methods developed for the analysis and integration of very large and diverse data sets generated by tandem MS measurements of tryptic peptides, we have now compiled a high-confidence human plasma proteome reference set with well over twice the identified proteins of previous high-confidence sets. It includes a hierarchy of protein identifications at different levels of redundancy following a clearly defined scheme, which we propose as a standard that can be applied to any proteomics data set to facilitate cross-proteome analyses. Further, to aid in development of blood-based diagnostics using techniques such as selected reaction monitoring, we provide a rough estimate of protein concentrations using spectral counting. We identified 20,433 distinct peptides, from which we inferred a highly nonredundant set of 1929 protein sequences at a false discovery rate of 1%. We have made this resource available via PeptideAtlas, a large, multiorganism, publicly accessible compendium of peptides identified in tandem MS experiments conducted by laboratories around the world.
Figures



References
-
- Putnam F. W. ed. (1975–1989) The Plasma Proteins, 2nd Ed., Academic Press, New York
-
- Anderson N. L., Anderson N. G. (2002) The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell Proteomics 1, 845–867 - PubMed
-
- Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. (2004) The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 - PubMed
-
- Omenn G. S., States D. J., Adamski M., Blackwell T. W., Menon R., Hermjakob H., Apweiler R., Haab B. B., Simpson R. J., Eddes J. S., Kapp E. A., Moritz R. L., Chan D. W., Rai A. J., Admon A., Aebersold R., Eng J., Hancock W. S., Hefta S. A., Meyer H., Paik Y. K., Yoo J. S., Ping P., Pounds J., Adkins J., Qian X., Wang R., Wasinger V., Wu C. Y., Zhao X., Zeng R., Archakov A., Tsugita A., Beer I., Pandey A., Pisano M., Andrews P., Tammen H., Speicher D. W., Hanash S. M. (2005) Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core data set of 3020 proteins and a publicly-available database. Proteomics 5, 3226–3245 - PubMed
-
- Omenn G. Editor (2006) Exploring the Human Plasma Proteome, Wiley-VCH, New York, NY
Publication types
MeSH terms
Substances
Grants and funding
- PM50 GMO7U547/PHS HHS/United States
- GM087221/GM/NIGMS NIH HHS/United States
- R01 GM087221/GM/NIGMS NIH HHS/United States
- PB0ES017885/ES/NIEHS NIH HHS/United States
- R44 HG004537/HG/NHGRI NIH HHS/United States
- P30 ES017885/ES/NIEHS NIH HHS/United States
- HG005805/HG/NHGRI NIH HHS/United States
- N01 HV028179/HL/NHLBI NIH HHS/United States
- U54 DA021519/DA/NIDA NIH HHS/United States
- 233226/ERC_/European Research Council/International
- RC2 HG005805/HG/NHGRI NIH HHS/United States
- R44HG004537/HG/NHGRI NIH HHS/United States
- N01-HV-28179/HV/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

