Tissue-specific differences in cytosine methylation and their association with differential gene expression in sorghum
- PMID: 21632971
- PMCID: PMC3149958
- DOI: 10.1104/pp.111.176842
Tissue-specific differences in cytosine methylation and their association with differential gene expression in sorghum
Abstract
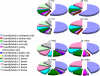
It has been well established that DNA cytosine methylation plays essential regulatory roles in imprinting gene expression in endosperm, and hence normal embryonic development, in the model plant Arabidopsis (Arabidopsis thaliana). Nonetheless, the developmental role of this epigenetic marker in cereal crops remains largely unexplored. Here, we report for sorghum (Sorghum bicolor) differences in relative cytosine methylation levels and patterns at 5'-CCGG sites in seven tissues (endosperm, embryo, leaf, root, young inflorescence, anther, and ovary), and characterize a set of tissue-specific differentially methylated regions (TDMRs). We found that the most enriched TDMRs in sorghum are specific for the endosperm and are generated concomitantly but imbalanced by decrease versus increase in cytosine methylation at multiple 5'-CCGG sites across the genome. This leads to more extensive demethylation in the endosperm than in other tissues, where TDMRs are mainly tissue nonspecific rather than specific to a particular tissue. Accordingly, relative to endosperm, the other six tissues showed grossly similar levels though distinct patterns of cytosine methylation, presumably as a result of a similar extent of concomitant decrease versus increase in cytosine methylation that occurred at variable genomic loci. All four tested TDMRs were validated by bisulfite genomic sequencing. Diverse sequences were found to underlie the TDMRs, including those encoding various known-function or predicted proteins, transposable elements, and those bearing homology to putative imprinted genes in maize (Zea mays). We further found that the expression pattern of at least some genic TDMRs was correlated with its tissue-specific methylation state, implicating a developmental role of DNA methylation in regulating tissue-specific or -preferential gene expression in sorghum.
Figures

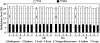




References
-
- Autran D, Huanca-Mamani W, Vielle-Calzada JP. (2005) Genomic imprinting in plants: the epigenetic version of an Oedipus complex. Curr Opin Plant Biol 8: 19–25 - PubMed
-
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE. (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13: 2212–2217 - PubMed
-
- Cao X, Jacobsen SE. (2002b) Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12: 1138–1144 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

