Irgm1 protects hematopoietic stem cells by negative regulation of IFN signaling
- PMID: 21633090
- PMCID: PMC3156044
- DOI: 10.1182/blood-2011-01-328682
Irgm1 protects hematopoietic stem cells by negative regulation of IFN signaling
Abstract
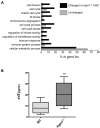
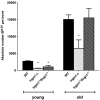
The IFN-inducible immunity-related p47 GTPase Irgm1 has been linked to Crohn disease as well as susceptibility to tuberculosis. Previously we demonstrated that HSC quiescence and function are aberrant in mice lacking Irgm1. To investigate the molecular basis for these defects, we conducted microarray expression profiling of Irgm1-deficient HSCs. Cell-cycle and IFN-response genes are up-regulated in Irgm1(-/-) HSCs, consistent with dysregulated IFN signaling. To test the hypothesis that Irgm1 normally down-regulates IFN signaling in HSCs, we generated Irgm1(-/-)Ifngr1(-/-) and Irgm1(-/-)Stat1(-/-) double-knockout animals. Strikingly, hyperproliferation, self-renewal, and autophagy defects in Irgm1(-/-) HSCs were normalized in double-knockout animals. These defects were also abolished in Irgm1(-/-)Irgm3(-/-) double-knockout animals, indicating that Irgm1 may regulate Irgm3 activity. Furthermore, the number of HSCs was reduced in aged Irgm1(-/-) animals, suggesting that negative feedback inhibition of IFN signaling by Irgm1 is necessary to prevent hyperproliferation and depletion of the stem cell compartment. Collectively, our results indicate that Irgm1 is a powerful negative regulator of IFN-dependent stimulation in HSCs, with an essential role in preserving HSC number and function. The deleterious effects of excessive IFN signaling may explain how hematologic abnormalities arise in patients with inflammatory conditions.
Figures



Comment in
-
Interferon and PV stem cells.Blood. 2011 Aug 11;118(6):1429-30. doi: 10.1182/blood-2011-06-359778. Blood. 2011. PMID: 21835958 No abstract available.
References
-
- Taylor G, Feng C, Sher A. Control of IFN-gamma–mediated host resistance to intracellular pathogens by immunity-related GTPases (p47 GTPases). Microbes Infect. 2007;9(14–15):1644–1651. - PubMed
-
- MacMicking JD, Taylor G, McKinney JD. Immune control of tuberculosis by IFN-gamma–inducible LRG-47. Science. 2003;302(5645):654–659. - PubMed
-
- Singh S. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313(5792):1438–1441. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK58192/DK/NIDDK NIH HHS/United States
- T32 HL092332/HL/NHLBI NIH HHS/United States
- P01 GM081627/GM/NIGMS NIH HHS/United States
- EB005173/EB/NIBIB NIH HHS/United States
- P30 CA125123/CA/NCI NIH HHS/United States
- R01 DK058192/DK/NIDDK NIH HHS/United States
- R01 AI057831/AI/NIAID NIH HHS/United States
- HL081007/HL/NHLBI NIH HHS/United States
- R01 EB005173/EB/NIBIB NIH HHS/United States
- K08 HL098898/HL/NHLBI NIH HHS/United States
- P50 CA126752/CA/NCI NIH HHS/United States
- P50CA126752/CA/NCI NIH HHS/United States
- F30 DK082107/DK/NIDDK NIH HHS/United States
- U54 HL081007/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

