Molecular Validation of PACE4 as a Target in Prostate Cancer
- PMID: 21633671
- PMCID: PMC3104696
- DOI: 10.1593/tlo.10295
Molecular Validation of PACE4 as a Target in Prostate Cancer
Abstract
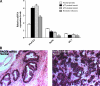
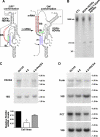
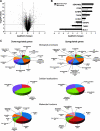
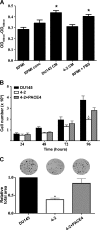
Prostate cancer remains the single most prevalent cancer in men. Standard therapies are still limited and include androgen ablation that initially causes tumor regression. However, tumor cells eventually relapse and develop into a hormone-refractory prostate cancer. One of the current challenges in this disease is to define new therapeutic targets, which have been virtually unchanged in the past 30 years. Recent studies have suggested that the family of enzymes known as the proprotein convertases (PCs) is involved in various types of cancers and their progression. The present study examined PC expression in prostate cancer and validates one PC, namely PACE4, as a target. The evidence includes the observed high expression of PACE4 in all different clinical stages of human prostate tumor tissues. Gene silencing studies targeting PACE4 in the DU145 prostate cancer cell line produced cells (cell line 4-2) with slower proliferation rates, reduced clonogenic activity, and inability to grow as xenografts in nude mice. Gene expression and proteomic profiling of the 4-2 cell line reveals an increased expression of known cancer-related genes (e.g., GJA1, CD44, IGFBP6) that are downregulated in prostate cancer. Similarly, cancer genes whose expression is decreased in the 4-2 cell line were upregulated in prostate cancer (e.g., MUC1, IL6). The direct role of PACE4 in prostate cancer is most likely through the upregulated processing of growth factors or through the aberrant processing of growth factors leading to sustained cancer progression, suggesting that PACE4 holds a central role in prostate cancer.
Figures


References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Bassi DE, Fu J, Lopez de Cicco R, Klein-Szanto AJ. Proprotein convertases: “master switches” in the regulation of tumor growth and progression. Mol Carcinog. 2005;44:151–161. - PubMed
-
- Blanchard A, Iwasiow B, Yarmill A, Fresnosa A, Silha J, Myal Y, Murphy LC, Chretien M, Seidah N, Shiu RP. Targeted production of proprotein convertase PC1 enhances mammary development and tumorigenesis in transgenic mice. Can J Physiol Pharmacol. 2009;87:831–838. - PubMed
-
- de Cicco RL, Bassi DE, Benavides F, Conti CJ, Klein-Szanto AJ. Inhibition of proprotein convertases: approaches to block squamous carcinoma development and progression. Mol Carcinog. 2007;46:654–659. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
