Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae
- PMID: 21635781
- PMCID: PMC3132168
- DOI: 10.1186/1471-2164-12-288
Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae
Abstract
Background: Emerging knowledge of the impact of small RNAs as important cellular regulators has prompted an explosion of small transcriptome sequencing projects. Although significant progress has been made towards small RNA discovery and biogenesis in higher eukaryotes and other model organisms, knowledge in simple eukaryotes such as filamentous fungi remains limited.
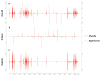
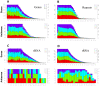
Results: Here, we used 454 pyrosequencing to present a detailed analysis of the small RNA transcriptome (~ 15 - 40 nucleotides in length) from mycelia and appressoria tissues of the rice blast fungal pathogen, Magnaporthe oryzae. Small RNAs mapped to numerous nuclear and mitochondrial genomic features including repetitive elements, tRNA loci, rRNAs, protein coding genes, snRNAs and intergenic regions. For most elements, small RNAs mapped primarily to the sense strand with the exception of repetitive elements to which small RNAs mapped in the sense and antisense orientation in near equal proportions. Inspection of the small RNAs revealed a preference for U and suppression of C at position 1, particularly for antisense mapping small RNAs. In the mycelia library, small RNAs of the size 18 - 23 nt were enriched for intergenic regions and repetitive elements. Small RNAs mapping to LTR retrotransposons were classified as LTR retrotransposon-siRNAs (LTR-siRNAs). Conversely, the appressoria library had a greater proportion of 28 - 35 nt small RNAs mapping to tRNA loci, and were classified as tRNA-derived RNA fragments (tRFs). LTR-siRNAs and tRFs were independently validated by 3' RACE PCR and northern blots, respectively.
Conclusions: Our findings suggest M. oryzae small RNAs differentially accumulate in vegetative and specialized-infection tissues and may play an active role in genome integrity and regulating growth and development.
Figures






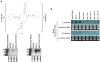

References
-
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 microRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

