Distinguishing arrhythmogenic right ventricular cardiomyopathy/dysplasia-associated mutations from background genetic noise
- PMID: 21636032
- PMCID: PMC6311127
- DOI: 10.1016/j.jacc.2010.12.036
Distinguishing arrhythmogenic right ventricular cardiomyopathy/dysplasia-associated mutations from background genetic noise
Abstract
Objectives: The aims of this study were to determine the spectrum and prevalence of "background genetic noise" in the arrhythmogenic right ventricular cardiomyopathy/dysplasia (ARVC) genetic test and to determine genetic associations that can guide the interpretation of a positive test result.
Background: ARVC is a potentially lethal genetic cardiovascular disorder characterized by myocyte loss and fibrofatty tissue replacement of the right ventricle. Genetic variation among the ARVC susceptibility genes has not been systematically examined, and little is known about the background noise associated with the ARVC genetic test.
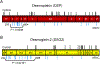
Methods: Using direct deoxyribonucleic acid sequencing, the coding exons/splice junctions of PKP2, DSP, DSG2, DSC2, and TMEM43 were genotyped for 93 probands diagnosed with ARVC from the Netherlands and 427 ostensibly healthy controls of various ethnicities. Eighty-two additional ARVC cases were obtained from published reports, and additional mutations were included from the ARVD/C Genetic Variants Database.
Results: The overall yield of mutations among ARVC cases was 58% versus 16% in controls. Radical mutations were hosted by 0.5% of control individuals versus 43% of ARVC cases, while 16% of controls hosted missense mutations versus a similar 21% of ARVC cases. Relative to controls, mutations in cases occurred more frequently in non-Caucasians, localized to the N-terminal regions of DSP and DSG2, and localized to highly conserved residues within PKP2 and DSG2.
Conclusions: This study is the first to comprehensively evaluate genetic variation in healthy controls for the ARVC susceptibility genes. Radical mutations are high-probability ARVC-associated mutations, whereas rare missense mutations should be interpreted in the context of race and ethnicity, mutation location, and sequence conservation.
Copyright © 2011 American College of Cardiology Foundation. Published by Elsevier Inc. All rights reserved.
Figures

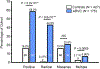


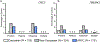

Comment in
-
Hearing the noise the challenges of human genome variation in genetic testing.J Am Coll Cardiol. 2011 Jun 7;57(23):2328-9. doi: 10.1016/j.jacc.2011.02.026. J Am Coll Cardiol. 2011. PMID: 21636033 Free PMC article. No abstract available.
References
-
- Peters S, Trümmel M, Meyners W. Prevalence of right ventricular dysplasia-cardiomyopathy in a non-referral hospital. Int. J. Cardiol 2004; 97:499–501. - PubMed
-
- Gemayel C, Pelliccia A, Thompson PD. Arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol. 2001; 38:1773–1781. - PubMed
-
- Dalal D, Molin LH, Piccini J, Tichnell C, James C, Bomma C, Prakasa K, Towbin JA, Marcus FI, Spevak PJ, Bluemke DA, Abraham T, Russell SD, Calkins H, Judge DP. Clinical Features of Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy Associated With Mutations in Plakophilin-2. Circulation. 2006; 113:1641–1649. - PubMed
-
- Sen-Chowdhry S, Syrris P, McKenna WJ. Role of Genetic Analysis in the Management of Patients With Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy. J Am Coll Cardiol. 2007; 50:1813–1821. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

