The kynurenine pathway modulates neurodegeneration in a Drosophila model of Huntington's disease
- PMID: 21636279
- PMCID: PMC3929356
- DOI: 10.1016/j.cub.2011.04.028
The kynurenine pathway modulates neurodegeneration in a Drosophila model of Huntington's disease
Abstract
Neuroactive metabolites of the kynurenine pathway (KP) of tryptophan degradation have been implicated in the pathophysiology of neurodegenerative disorders, including Huntington's disease (HD) [1]. A central hallmark of HD is neurodegeneration caused by a polyglutamine expansion in the huntingtin (htt) protein [2]. Here we exploit a transgenic Drosophila melanogaster model of HD to interrogate the therapeutic potential of KP manipulation. We observe that genetic and pharmacological inhibition of kynurenine 3-monooxygenase (KMO) increases levels of the neuroprotective metabolite kynurenic acid (KYNA) relative to the neurotoxic metabolite 3-hydroxykynurenine (3-HK) and ameliorates neurodegeneration. We also find that genetic inhibition of tryptophan 2,3-dioxygenase (TDO), the first and rate-limiting step in the pathway, leads to a similar neuroprotective shift toward KYNA synthesis. Importantly, we demonstrate that the feeding of KYNA and 3-HK to HD model flies directly modulates neurodegeneration, underscoring the causative nature of these metabolites. This study provides the first genetic evidence that inhibition of KMO and TDO activity protects against neurodegenerative disease in an animal model, indicating that strategies targeted at two key points within the KP may have therapeutic relevance in HD, and possibly other neurodegenerative disorders.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

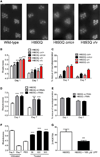
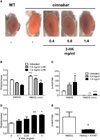

Comment in
-
Neurodegenerative disease: the kynurenine pathway--promising new targets and therapies for neurodegenerative disease.Nat Rev Neurol. 2011 Jul 26;7(8):417. doi: 10.1038/nrneurol.2011.102. Nat Rev Neurol. 2011. PMID: 21788979 No abstract available.
-
Neurodegenerative disorders: restoring the balance.Nat Rev Drug Discov. 2011 Aug 1;10(8):576. doi: 10.1038/nrd3521. Nat Rev Drug Discov. 2011. PMID: 21804592 No abstract available.
References
-
- Thevandavakkam MA, Schwarcz R, Muchowski PJ, Giorgini F. Targeting kynurenine 3-monooxygenase (KMO): implications for therapy in Huntington’s disease. CNS Neurol. Disord. Drug Targets. 2010;9:791–800. - PubMed
-
- The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. - PubMed
-
- Ishii T, Iwahashi H, Sugata R, Kido R. Formation of hydroxanthommatin-derived radical in the oxidation of 3-hydroxykynurenine. Arch. Biochem. Biophys. 1992;294:616–622. - PubMed
-
- Hiraku Y, Inoue S, Oikawa S, Yamamoto K, Tada S, Nishino K, Kawanishi S. Metal-mediated oxidative damage to cellular and isolated DNA by certain tryptophan metabolites. Carcinogenesis. 1995;16:349–356. - PubMed
-
- Stone TW, Perkins MN. Quinolinic acid: a potent endogenous excitant at amino acid receptors in CNS. Eur. J. Pharmacol. 1981;72:411–412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

