Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis
- PMID: 21636745
- PMCID: PMC3235694
- DOI: 10.1126/science.1203285
Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis
Abstract
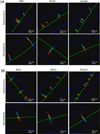
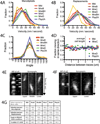
Rod-shaped bacteria elongate by the action of cell wall synthesis complexes linked to underlying dynamic MreB filaments. To understand how the movements of these filaments relate to cell wall synthesis, we characterized the dynamics of MreB and the cell wall elongation machinery using high-precision particle tracking in Bacillus subtilis. We found that MreB and the elongation machinery moved circumferentially around the cell, perpendicular to its length, with nearby synthesis complexes and MreB filaments moving independently in both directions. Inhibition of cell wall synthesis by various methods blocked the movement of MreB. Thus, bacteria elongate by the uncoordinated, circumferential movements of synthetic complexes that insert radial hoops of new peptidoglycan during their transit, possibly driving the motion of the underlying MreB filaments.
Figures


Comment in
-
Bacterial physiology: MreB takes a back seat.Nat Rev Microbiol. 2011 Jul 4;9(8):560-1. doi: 10.1038/nrmicro2621. Nat Rev Microbiol. 2011. PMID: 21725336 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

