Pre-microRNA and mature microRNA in human mitochondria
- PMID: 21637849
- PMCID: PMC3102686
- DOI: 10.1371/journal.pone.0020220
Pre-microRNA and mature microRNA in human mitochondria
Abstract
Background: Because of the central functions of the mitochondria in providing metabolic energy and initiating apoptosis on one hand and the role that microRNA (miRNA) play in gene expression, we hypothesized that some miRNA could be present in the mitochondria for post-transcriptomic regulation by RNA interference. We intend to identify miRNA localized in the mitochondria isolated from human skeletal primary muscular cells.
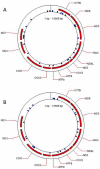
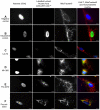
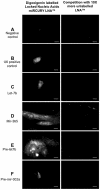
Methodology/principal findings: To investigate the potential origin of mitochondrial miRNA, we in-silico searched for microRNA candidates in the mtDNA. Twenty five human pre-miRNA and 33 miRNA aligments (E-value<0.1) were found in the reference mitochondrial sequence and some of the best candidates were chosen for a co-localization test. In situ hybridization of pre-mir-302a, pre-let-7b and mir-365, using specific labelled locked nucleic acids and confocal microscopy, demonstrated that these miRNA were localized in mitochondria of human myoblasts. Total RNA was extracted from enriched mitochondria isolated by an immunomagnetic method from a culture of human myotubes. The detection of 742 human miRNA (miRBase) were monitored by RT-qPCR at three increasing mtRNA inputs. Forty six miRNA were significantly expressed (2(nd) derivative method Cp>35) for the smallest RNA input concentration and 204 miRNA for the maximum RNA input concentration. In silico analysis predicted 80 putative miRNA target sites in the mitochondrial genome (E-value<0.05).
Conclusions/significance: The present study experimentally demonstrated for the first time the presence of pre-miRNA and miRNA in the human mitochondria isolated from skeletal muscular cells. A set of miRNA were significantly detected in mitochondria fraction. The origin of these pre-miRNA and miRNA should be further investigate to determine if they are imported from the cytosol and/or if they are partially processed in the mitochondria.
Conflict of interest statement
Figures



References
-
- Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410:103–123. - PubMed
-
- Mokranjac D, Neupert W. Protein import into isolated mitochondria. Methods Mol Biol. 2007;372:277–286. - PubMed
-
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

