BEL/Pao retrotransposons in metazoan genomes
- PMID: 21639932
- PMCID: PMC3118150
- DOI: 10.1186/1471-2148-11-154
BEL/Pao retrotransposons in metazoan genomes
Abstract
Background: Long terminal repeat (LTR) retrotransposons are a widespread kind of transposable element present in eukaryotic genomes. They are a major factor in genome evolution due to their ability to create large scale mutations and genome rearrangements. Compared to other transposable elements, little attention has been paid to elements belonging to the metazoan BEL/Pao subclass of LTR retrotransposons. No comprehensive characterization of these elements is available so far. The aim of this study was to describe all BEL/Pao elements in a set of 62 sequenced metazoan genomes, and to analyze their phylogenetic relationship.
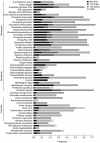
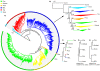
Results: We identified a total of 7,861 BEL/Pao elements in 53 of our 62 study genomes. We identified BEL/Pao elements in 20 genomes where such elements had not been found so far. Our analysis shows that BEL/Pao elements are the second-most abundant class of LTR retrotransposons in the genomes we study, more abundant than Ty1/Copia elements, and second only to Ty3/Gypsy elements. They occur in multiple phyla, including basal metazoan phyla, suggesting that BEL/Pao elements arose early in animal evolution. We confirm findings from previous studies that BEL/Pao elements do not occur in mammals. The elements we found can be grouped into more than 1725 families, 1623 of which are new, previously unknown families. These families fall into seven superfamilies, only five of which have been characterized so far. One new superfamily is a major subdivision of the Pao superfamily which we propose to call Dan, because it is restricted to the genome of the zebrafish Danio rerio. The other new superfamily comprises 83 elements and is restricted to lower aquatic eumetazoans. We propose to call this superfamily Flow. BEL/Pao elements do not show any signs of recent horizontal gene transfer between distantly related species.
Conclusions: In sum, our analysis identifies thousands of new BEL/Pao elements and provides new insights into their distribution, abundance, and evolution.
Figures


References
-
- Craig NL, Craigie R, Gellert M, Lambowitz AM, (Eds) Mobile DNA II. Washington: ASM Press; 2002.
-
- Kidwell MG, Lisch DR. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution Int J Org Evolution. 2001;55:1–24. http://www.jstor.org/pss/2640685 - PubMed
-
- Ganko EW, Bhattacharjee V, Schliekelman P, McDonald JF. Evidence for the contribution of LTR retrotransposons to C. elegans gene evolution. Mol Biol Evol. 2003;20(11):1925–1931. doi: 10.1093/molbev/msg200. http://dx.doi.org/10.1093/molbev/msg200 - DOI - DOI - PubMed
-
- Eickbush TH, Malik HS. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editor. Herndon: ASM Press; 2002. Origins and evolution of retrotransposons; pp. 1111–1144.
-
- Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9(10):3353–3362. http://www.ncbi.nlm.nih.gov/pubmed/1698615?dopt=Abstract - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

