The lamin protein family
- PMID: 21639948
- PMCID: PMC3219962
- DOI: 10.1186/gb-2011-12-5-222
The lamin protein family
Abstract
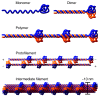
The lamins are the major architectural proteins of the animal cell nucleus. Lamins line the inside of the nuclear membrane, where they provide a platform for the binding of proteins and chromatin and confer mechanical stability. They have been implicated in a wide range of nuclear functions, including higher-order genome organization, chromatin regulation, transcription, DNA replication and DNA repair. The lamins are members of the intermediate filament (IF) family of proteins, which constitute a major component of the cytoskeleton. Lamins are the only nuclear IFs and are the ancestral founders of the IF protein superfamily. Lamins polymerize into fibers forming a complex protein meshwork in vivo and, like all IF proteins, have a tripartite structure with two globular head and tail domains flanking a central α-helical rod domain, which supports the formation of higher-order polymers. Mutations in lamins cause a large number of diverse human diseases, collectively known as the laminopathies, underscoring their functional importance.
Figures





References
-
- Lin F, Worman HJ. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J Biol Chem. 1993;268:16321–16326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

