Propulsive forces on the flagellum during locomotion of Chlamydomonas reinhardtii
- PMID: 21641317
- PMCID: PMC3117176
- DOI: 10.1016/j.bpj.2011.05.001
Propulsive forces on the flagellum during locomotion of Chlamydomonas reinhardtii
Abstract
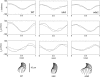
The distributed propulsive forces exerted on the flagellum of the swimming alga Chlamydomonas reinhardtii by surrounding fluid were estimated from experimental image data. Images of uniflagellate mutant Chlamydomonas cells were obtained at 350 frames/s with 125-nm spatial resolution, and the motion of the cell body and the flagellum were analyzed in the context of low-Reynolds-number fluid mechanics. Wild-type uniflagellate cells, as well as uniflagellate cells lacking inner dynein arms (ida3) or outer dynein arms (oda2) were studied. Ida3 cells exhibit stunted flagellar waveforms, whereas oda2 cells beat with lower frequency. Image registration and sorting algorithms provided high-resolution estimates of the motion of the cell body, as well as detailed kinematics of the flagellum. The swimming cell was modeled as an ellipsoid in Stokes flow, propelled by viscous forces on the flagellum. The normal and tangential components of force on the flagellum (f(N) and f(T)) were related by resistive coefficients (C(N) and C(T)) to the corresponding components of velocity (V(N) and V(T)).The values of these coefficients were estimated by satisfying equilibrium requirements for force and torque on the cell. The estimated values of the resistive coefficients are consistent among all three genotypes and similar to theoretical predictions.
Copyright © 2011 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures








References
-
- Lindemann C.B. A model of flagellar and ciliary functioning which uses the forces transverse to the axoneme as the regulator of dynein activation. Cell Motil. Cytoskeleton. 1994;29:141–154. - PubMed
-
- Lindemann C.B. Testing the geometric clutch hypothesis. Biol. Cell. 2004;96:681–690. - PubMed
-
- Huang B., Ramanis Z., Luck D.J. Uniflagellar mutants of Chlamydomonas: evidence for the role of basal bodies in transmission of positional information. Cell. 1982;29:745–753. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

