How dormant origins promote complete genome replication
- PMID: 21641805
- PMCID: PMC3329722
- DOI: 10.1016/j.tibs.2011.05.002
How dormant origins promote complete genome replication
Abstract
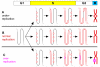
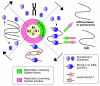
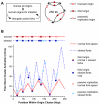
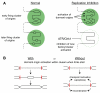
Many replication origins that are licensed by loading MCM2-7 complexes in G1 are not normally used. Activation of these dormant origins during S phase provides a first line of defence for the genome if replication is inhibited. When replication forks fail, dormant origins are activated within regions of the genome currently engaged in replication. At the same time, DNA damage-response kinases activated by the stalled forks preferentially suppress the assembly of new replication factories, thereby ensuring that chromosomal regions experiencing replicative stress complete synthesis before new regions of the genome are replicated. Mice expressing reduced levels of MCM2-7 have fewer dormant origins, are cancer-prone and are genetically unstable, demonstrating the importance of dormant origins for preserving genome integrity. We review the function of dormant origins, the molecular mechanism of their regulation and their physiological implications.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

References
-
- Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

