Synthesis and characterization of chimeric proteins based on cellulase and xylanase from an insect gut bacterium
- PMID: 21642416
- PMCID: PMC3147366
- DOI: 10.1128/AEM.02808-10
Synthesis and characterization of chimeric proteins based on cellulase and xylanase from an insect gut bacterium
Abstract
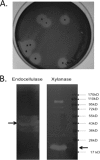
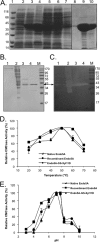
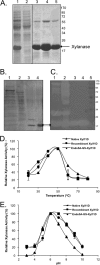
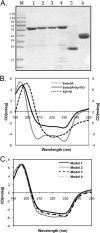
Insects living on wood and plants harbor a large variety of bacterial flora in their guts for degrading biomass. We isolated a Paenibacillus strain, designated ICGEB2008, from the gut of a cotton bollworm on the basis of its ability to secrete a variety of plant-hydrolyzing enzymes. In this study, we cloned, expressed, and characterized two enzymes, β-1,4-endoglucanase (Endo5A) and β-1,4-endoxylanase (Xyl11D), from the ICGEB2008 strain and synthesized recombinant bifunctional enzymes based on Endo5A and Xyl11D. The gene encoding Endo5A was obtained from the genome of the ICGEB2008 strain by shotgun cloning. The gene encoding Xyl11D was obtained using primers for conserved xylanase sequences, which were identified by aligning xylanase sequences in other species of Paenibacillus. Endo5A and Xyl11D were overexpressed in Escherichia coli, and their optimal activities were characterized. Both Endo5A and Xyl11D exhibited maximum specific activity at 50°C and pH 6 to 7. To take advantage of this feature, we constructed four bifunctional chimeric models of Endo5A and Xyl11D by fusing the encoding genes either end to end or through a glycine-serine (GS) linker. We predicted three-dimensional structures of the four models using the I-TASSER server and analyzed their secondary structures using circular dichroism (CD) spectroscopy. The chimeric model Endo5A-GS-Xyl11D, in which a linker separated the two enzymes, yielded the highest C-score on the I-TASSER server, exhibited secondary structure properties closest to the native enzymes, and demonstrated 1.6-fold and 2.3-fold higher enzyme activity than Endo5A and Xyl11D, respectively. This bifunctional enzyme could be effective for hydrolyzing plant biomass owing to its broad substrate range.
Figures
References
-
- An J. M., et al. 2005. Evaluation of a novel bifunctional xylanase-cellulase constructed by gene fusion. Enzyme Microb. Technol. 36:989–995
-
- Beliën T., Van Campenhout S., Robben J., Volckaert G. 2006. Microbial endoxylanases: effective weapons to breach the plant cell-wall barrier or, rather, triggers of plant defense systems? Mol. Plant Microbe Interact. 19:1072–1081 - PubMed
-
- Cho K. M., et al. 2008. Cloning of two cellulase genes from endophytic Paenibacillus polymyxa GS01 and comparison with cel 44C-man 26A. J. Basic Microbiol. 48:464–472 - PubMed
-
- Dashtban M., Maki M., Leung K. T., Mao C., Qin W. 2010. Cellulase activities in biomass conversion: measurement methods and comparison. Crit. Rev. Biotechnol. 30:302–309 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

