Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation
- PMID: 21642539
- PMCID: PMC3119735
- DOI: 10.4049/jimmunol.1100750
Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation
Abstract
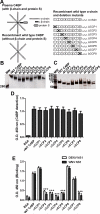
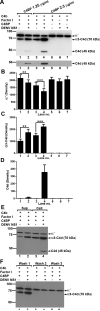
The complement system plays a pivotal protective role in the innate immune response to many pathogens including flaviviruses. Flavivirus nonstructural protein 1 (NS1) is a secreted nonstructural glycoprotein that accumulates in plasma to high levels and is displayed on the surface of infected cells but absent from viral particles. Previous work has defined an immune evasion role of flavivirus NS1 in limiting complement activation by forming a complex with C1s and C4 to promote cleavage of C4 to C4b. In this study, we demonstrate a second mechanism, also involving C4 and its active fragment C4b, by which NS1 antagonizes complement activation. Dengue, West Nile, or yellow fever virus NS1 directly associated with C4b binding protein (C4BP), a complement regulatory plasma protein that attenuates the classical and lectin pathways. Soluble NS1 recruited C4BP to inactivate C4b in solution and on the plasma membrane. Mapping studies revealed that the interaction sites of NS1 on C4BP partially overlap with the C4b binding sites. Together, these studies further define the immune evasion potential of NS1 in reducing the functional capacity of C4 in complement activation and control of flavivirus infection.
Figures
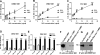


References
-
- de Cordoba SR, Garcia OC, Sanchez-Corral P. C4b-binding protein. In: Morley BJ, Walport MJ, editors. The complement facts book. Academic Press; London: 2000. pp. 161–167.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

