The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma
- PMID: 21642991
- PMCID: PMC4643098
- DOI: 10.1038/ng.855
The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma
Abstract
Malignant pleural mesotheliomas (MPMs) often show CDKN2A and NF2 inactivation, but other highly recurrent mutations have not been described. To identify additional driver genes, we used an integrated genomic analysis of 53 MPM tumor samples to guide a focused sequencing effort that uncovered somatic inactivating mutations in BAP1 in 23% of MPMs. The BAP1 nuclear deubiquitinase is known to target histones (together with ASXL1 as a Polycomb repressor subunit) and the HCF1 transcriptional co-factor, and we show that BAP1 knockdown in MPM cell lines affects E2F and Polycomb target genes. These findings implicate transcriptional deregulation in the pathogenesis of MPM.
Figures


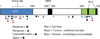

References
-
- Illei PB, Rusch VW, Zakowski MF, Ladanyi M. Homozygous deletion of CDKN2A and codeletion of the methylthioadenosine phosphorylase gene in the majority of pleural mesotheliomas. Clin. Cancer. Res. 2003;9:2108–2113. - PubMed
-
- Thurneysen C, et al. Functional inactivation of NF2/merlin in human mesothelioma. Lung Cancer. 2009;64:140–147. - PubMed
-
- Lu YY, Jhanwar SC, Cheng JQ, Testa JR. Deletion mapping of the short arm of chromosome 3 in human malignant mesothelioma. Genes Chromosom. Cancer. 1994;9:76–80. - PubMed
-
- López-Ríos F, et al. Global gene expression profiling of pleural mesotheliomas: overexpression of aurora kinases and p16/CDKN2A deletion as prognostic factors and critical evaluation of microarray-based prognostic prediction. Cancer. Res. 2006;66:2970–2979. - PubMed
-
- Jensen DE, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

