Visualizing fitness landscapes
- PMID: 21644947
- PMCID: PMC3668694
- DOI: 10.1111/j.1558-5646.2011.01236.x
Visualizing fitness landscapes
Abstract
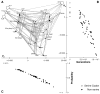
Fitness landscapes are a classical concept for thinking about the relationship between genotype and fitness. However, because the space of genotypes is typically high-dimensional, the structure of fitness landscapes can be difficult to understand and the heuristic approach of thinking about fitness landscapes as low-dimensional, continuous surfaces may be misleading. Here, I present a rigorous method for creating low-dimensional representations of fitness landscapes. The basic idea is to plot the genotypes in a manner that reflects the ease or difficulty of evolving from one genotype to another. Such a layout can be constructed using the eigenvectors of the transition matrix describing the evolution of a population on the fitness landscape when mutation is weak. In addition, the eigendecomposition of this transition matrix provides a new, high-level view of evolution on a fitness landscape. I demonstrate these techniques by visualizing the fitness landscape for selection for the amino acid serine and by visualizing a neutral network derived from the RNA secondary structure genotype-phenotype map.
© 2011 The Author(s). Evolution© 2011 The Society for the Study of Evolution.
Figures


References
-
- Ashlock D, Schonfeld J. Proceedings of the 2005 Congress on Evolutionary Computation. Vol. 3. Springer; 2005. Nonlinear projection for the display of high dimensional distance data; pp. 2776–2783.
-
- Barton NH, Coe JB. On the application of statistical physics to evolutionary biology. J Theor Biol. 2009;259:317–324. - PubMed
-
- Belkin M, Niyogi P. Laplacian eigenmaps for dimensionality reduction and data representation. Neural Comput. 2003;15:1373–1396.
