Dynamics may significantly influence the estimation of interatomic distances in biomolecular X-ray structures
- PMID: 21645520
- PMCID: PMC3171141
- DOI: 10.1016/j.jmb.2011.05.033
Dynamics may significantly influence the estimation of interatomic distances in biomolecular X-ray structures
Abstract
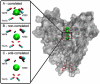
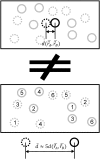
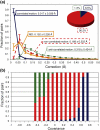
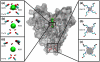
Atomic positions obtained by X-ray crystallography are time and space averages over many molecules in the crystal. Importantly, interatomic distances, calculated between such average positions and frequently used in structural and mechanistic analyses, can be substantially different from the more appropriate time-average and ensemble-average interatomic distances. Using crystallographic B-factors, one can deduce corrections, which have so far been applied exclusively to small molecules, to obtain correct average distances as a function of the type of atomic motion. Here, using 4774 high-quality protein X-ray structures, we study the significance of such corrections for different types of atomic motion. Importantly, we show that for distances shorter than 5 Å, corrections greater than 0.5 Å may apply, especially for noncorrelated or anticorrelated motion. For example, 14% of the studied structures have at least one pair of atoms with a correction of ≥0.5 Å in the case of noncorrelated motion. Using molecular dynamics simulations of villin headpiece, ubiquitin, and SH3 domain unit cells, we demonstrate that the majority of average interatomic distances in these proteins agree with noncorrelated corrections, suggesting that such deviations may be truly relevant. Importantly, we demonstrate that the corrections do not significantly affect stereochemistry and the overall quality of final refined X-ray structures, but can provide marked improvements in starting unrefined models obtained from low-resolution X-ray data. Finally, we illustrate the potential mechanistic and biological significance of the calculated corrections for KcsA ion channel and show that they provide indirect evidence that motions in its selectivity filter are highly correlated.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures


References
-
- Kruschel D., Zagrovic B. Conformational averaging in structural biology: issues, challenges and computational solutions. Mol. Biosyst. 2009;5:1606–1616. - PubMed
-
- Busing W.R., Levy H.A. The effect of thermal motion on estimation of bond lengths from diffraction measurements. Acta Crystallogr. 1964;17:142–146.
-
- Watkin D. Structure refinement: some background theory and practical strategies. J. Appl. Crystallogr. 2008;41:491–522.
-
- Bürgi H.B. Motion and disorder in crystal structure analysis: measuring and distinguishing them. Annu. Rev. Phys. Chem. 2000;51:275–296. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

