Domain orientation in the N-Terminal PDZ tandem from PSD-95 is maintained in the full-length protein
- PMID: 21645852
- PMCID: PMC3116789
- DOI: 10.1016/j.str.2011.02.017
Domain orientation in the N-Terminal PDZ tandem from PSD-95 is maintained in the full-length protein
Abstract
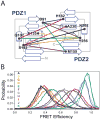
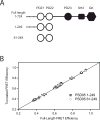
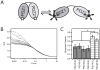
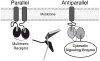
Tandem PDZ domains have been suggested to form structurally independent supramodules. However, dissimilarity between crystallography and NMR models emphasize their malleable conformation. Studies in full-length scaffold proteins are needed to examine the effect of tertiary interactions within their native context. Using single-molecule fluorescence to characterize the N-terminal PDZ tandem in PSD-95, we provide the first direct evidence that PDZ tandems can be structurally independent within a full-length scaffold protein. Molecular refinement using our data converged on a single structure with an antiparallel alignment of the ligand-binding sites. Devoid of interaction partners, single-molecule conditions captured PSD-95 in its unbound, ground state. Interactions between PDZ domains could not be detected while fluctuation correlation spectroscopy showed that other conformations are dynamically sampled. We conclude that ultra-weak interactions stabilize the conformation providing a "low-relief" energy landscape that allows the domain orientation to be flipped by environmental interactions.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures



References
-
- Bhatnagar J, Freed JH, Crane BR. Rigid body refinement of protein complexes with long-range distance restraints from pulsed dipolar ESR. Methods Enzymol. 2007;423:117–133. - PubMed
-
- Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem. 2006;75:655–680. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

