UV stalled replication forks restart by re-priming in human fibroblasts
- PMID: 21646340
- PMCID: PMC3167624
- DOI: 10.1093/nar/gkr420
UV stalled replication forks restart by re-priming in human fibroblasts
Abstract
Restarting stalled replication forks is vital to avoid fatal replication errors. Previously, it was demonstrated that hydroxyurea-stalled replication forks rescue replication either by an active restart mechanism or by new origin firing. To our surprise, using the DNA fibre assay, we only detect a slightly reduced fork speed on a UV-damaged template during the first hour after UV exposure, and no evidence for persistent replication fork arrest. Interestingly, no evidence for persistent UV-induced fork stalling was observed even in translesion synthesis defective, Polη(mut) cells. In contrast, using an assay to measure DNA molecule elongation at the fork, we observe that continuous DNA elongation is severely blocked by UV irradiation, particularly in UV-damaged Polη(mut) cells. In conclusion, our data suggest that UV-blocked replication forks restart effectively through re-priming past the lesion, leaving only a small gap opposite the lesion. This allows continuation of replication on damaged DNA. If left unfilled, the gaps may collapse into DNA double-strand breaks that are repaired by a recombination pathway, similar to the fate of replication forks collapsed after hydroxyurea treatment.
Figures

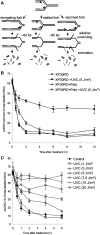
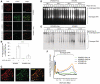

References
-
- Hekmat-Nejad M, You Z, Yee MC, Newport JW, Cimprich KA. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr. Biol. 2000;10:1565–1573. - PubMed
-
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

